Word cloud of the abstracts that follow.
56.
Tsampazi, V. & Glykos*, N.M. (2026),
"Quantifying the uncertainty of molecular dynamics simulations : Good-Turing statistics revisited",
arXiv:2601.00618 [q-bio.QM]
Electronic reprint (1.6 MBytes) : Local copy, or directly from arXiv:2601.00618.

 We have previously shown that Good-Turing statistics can be applied to molecular
dynamics trajectories to estimate the probability of observing completely new
(thus far unobserved) biomolecular structures, and showed that the method is
stable, dependable and its predictions verifiable. The major problem with that
initial algorithm was the requirement for calculating and storing in memory the
two-dimensional RMSD matrix of the currently available trajectory. This
requirement precluded the application of the method to very long simulations.
Here we describe a new variant of the Good-Turing algorithm whose memory
requirements scale linearly with the number of structures in the trajectory,
making it suitable even for extremely long simulations. We show that the new
method gives essentially identical results with the older implementation, and
present results obtained from trajectories containing up to 22 million
structures. A computer program implementing the new algorithm is available from
standard repositories.
We have previously shown that Good-Turing statistics can be applied to molecular
dynamics trajectories to estimate the probability of observing completely new
(thus far unobserved) biomolecular structures, and showed that the method is
stable, dependable and its predictions verifiable. The major problem with that
initial algorithm was the requirement for calculating and storing in memory the
two-dimensional RMSD matrix of the currently available trajectory. This
requirement precluded the application of the method to very long simulations.
Here we describe a new variant of the Good-Turing algorithm whose memory
requirements scale linearly with the number of structures in the trajectory,
making it suitable even for extremely long simulations. We show that the new
method gives essentially identical results with the older implementation, and
present results obtained from trajectories containing up to 22 million
structures. A computer program implementing the new algorithm is available from
standard repositories.
55. Touliopoulos, S. & Glykos*, N.M. (2026), "Atomic density distributions in proteins: structural and functional implications", J. Mol. Graph. Model., 142, 109228.
Electronic reprint (9.0 MBytes) : Local copy, or directly from J. Mol. Graph. Model.. See also the Supporting information file.

 Atomic packing is an important metric for characterizing protein structures, as
it significantly influences various features including the stability, the rate
of evolution and the functional roles of proteins. Packing in protein structures
is a measure of the overall proximity between the proteins’ atoms and it can
vary notably among different structures. However, even single domain proteins do
not exhibit uniform packing throughout their structure. Protein cores in the
interior tend to be more tightly packed compared to the protein surface and the
presence of cavities and voids can disrupt that internal tight packing too.
Many different methods have been used to measure the quality of packing in
proteins, identify factors that influence it, and their possible implications.
In this work, we examine atomic density distributions derived from 21,255
non-redundant protein structures and show that statistically significant
differences between those distributions are present. The biomolecular assembly
unit was chosen as a representative for these structures. Addition of hydrogen
atoms and solvation was also performed to emulate a faithful representation of
the structures in vitro.
Several protein structures deviate significantly and systematically from the
average packing behavior. Hierarchical clustering indicated that there are
groups of structures with similar atomic density distributions. Search for
common features and patterns in these clusters showed that some of them include
proteins with characteristic structures such as coiled-coils and cytochromes.
Certain classification families such as hydrolases and transferases have also a
preference to appear more frequently in dense and loosely-packed clusters
respectively.
Regarding factors influencing packing, our results support knowledge that larger
structures have a smaller range in their density values, but tend to be more
loosely packed, compared to smaller proteins. We also used indicators, like
crystallographic water molecules abundance and B-factors as estimates of the
stability of the structures to reveal its relationship with packing.
Atomic packing is an important metric for characterizing protein structures, as
it significantly influences various features including the stability, the rate
of evolution and the functional roles of proteins. Packing in protein structures
is a measure of the overall proximity between the proteins’ atoms and it can
vary notably among different structures. However, even single domain proteins do
not exhibit uniform packing throughout their structure. Protein cores in the
interior tend to be more tightly packed compared to the protein surface and the
presence of cavities and voids can disrupt that internal tight packing too.
Many different methods have been used to measure the quality of packing in
proteins, identify factors that influence it, and their possible implications.
In this work, we examine atomic density distributions derived from 21,255
non-redundant protein structures and show that statistically significant
differences between those distributions are present. The biomolecular assembly
unit was chosen as a representative for these structures. Addition of hydrogen
atoms and solvation was also performed to emulate a faithful representation of
the structures in vitro.
Several protein structures deviate significantly and systematically from the
average packing behavior. Hierarchical clustering indicated that there are
groups of structures with similar atomic density distributions. Search for
common features and patterns in these clusters showed that some of them include
proteins with characteristic structures such as coiled-coils and cytochromes.
Certain classification families such as hydrolases and transferases have also a
preference to appear more frequently in dense and loosely-packed clusters
respectively.
Regarding factors influencing packing, our results support knowledge that larger
structures have a smaller range in their density values, but tend to be more
loosely packed, compared to smaller proteins. We also used indicators, like
crystallographic water molecules abundance and B-factors as estimates of the
stability of the structures to reveal its relationship with packing.
54. Vouzina, O.-D., Tafanidis, A. & Glykos*, N.M. (2024), "The curious case of A31P, a topology-switching mutant of the Repressor of Primer protein : A molecular dynamics study of its folding and misfolding", J. Chem. Inf. Model., 64, 6081-6091.
Electronic reprint (6.3 MBytes) : Local copy, or directly from J. Chem. Inf. Model.. See also the Supporting information file.

 The effect of mutations on protein structures is usually rather localized and
minor. Finding a mutation that can single-handedly change the fold and/or
topology of a protein structure is a rare exception. The A31P mutant of the
homodimeric Repressor of Primer (Rop) protein is one such exception: This single
mutation -and as demonstrated by two independent crystal structure
determinations- can convert the canonical (left-handed/all-antiparallel)
4-alpha-helical bundle of Rop, to a new form (right-handed/mixed parallel and
antiparallel bundle) displaying a previously unobserved 'bisecting U' topology.
The main problem with understanding the dramatic effect of this mutation on the
folding of Rop is to understand its very existence : Most computational methods
appear to agree that the mutation should have had no appreciable effect, with
the majority of energy minimization methods and protein structure prediction
protocols indicating that this mutation is fully consistent with the native Rop
structure, requiring only a local and minor change at the mutation site. Here we
use two long (10 us each) molecular dynamics simulations to compare the
stability and dynamics of the native Rop versus a hypothetical structure that is
identical with the native Rop but is carrying this single Alanine-31 to Proline
mutation. Comparative analysis of the two trajectories convincingly shows that
in contrast to the indications from energy minimization -but in agreement with
the experimental data-, this hypothetical native-like A31P structure is
unstable, with its turn regions almost completely unfolding, even under the
relatively mild 320K NpT simulations that we have used for this study. We
discuss the implication of these findings for the folding of the A31P mutant,
especially with respect to the proposed model of a double-funneled energy
landscape.
The effect of mutations on protein structures is usually rather localized and
minor. Finding a mutation that can single-handedly change the fold and/or
topology of a protein structure is a rare exception. The A31P mutant of the
homodimeric Repressor of Primer (Rop) protein is one such exception: This single
mutation -and as demonstrated by two independent crystal structure
determinations- can convert the canonical (left-handed/all-antiparallel)
4-alpha-helical bundle of Rop, to a new form (right-handed/mixed parallel and
antiparallel bundle) displaying a previously unobserved 'bisecting U' topology.
The main problem with understanding the dramatic effect of this mutation on the
folding of Rop is to understand its very existence : Most computational methods
appear to agree that the mutation should have had no appreciable effect, with
the majority of energy minimization methods and protein structure prediction
protocols indicating that this mutation is fully consistent with the native Rop
structure, requiring only a local and minor change at the mutation site. Here we
use two long (10 us each) molecular dynamics simulations to compare the
stability and dynamics of the native Rop versus a hypothetical structure that is
identical with the native Rop but is carrying this single Alanine-31 to Proline
mutation. Comparative analysis of the two trajectories convincingly shows that
in contrast to the indications from energy minimization -but in agreement with
the experimental data-, this hypothetical native-like A31P structure is
unstable, with its turn regions almost completely unfolding, even under the
relatively mild 320K NpT simulations that we have used for this study. We
discuss the implication of these findings for the folding of the A31P mutant,
especially with respect to the proposed model of a double-funneled energy
landscape.
53. Kolocouris*, A., Arkin, I. & Glykos*, N.M. (2022), "A proof-of-concept study of the secondary structure of influenza A, B M2 and MERS- and SARS-CoV E transmembrane peptides using folding molecular dynamics simulations in a membrane mimetic solvent", Phys. Chem. Chem. Phys., 24, 25391-25402.
Electronic reprint (1.8 MBytes) : Local copy, or directly from Phys. Chem. Chem. Phys., Copyright © Royal Society of Chemistry.

 We have carried out a proof-of-concept molecular dynamics (MD) simulation
with adaptive tempering in a membrane mimetic environment to study the folding
of single-pass membrane peptides. We tested the influenza A M2 viroporin,
influenza B M2 viroporin, and protein E from coronaviruses MERS-Cov-2 and
SARS-CoV-2 peptides with known experimental secondary structures in membrane
bilayers. The two influenza-derived peptides are significantly different in the
peptide sequence and secondary structure and more polar than the two
coronavirus-derived peptides. Through a total of more than 50 μs of simulation
time that could be accomplished in trifluoroethanol (TFE), as a membrane model,
we characterized comparatively the folding behavior, helical stability, and
helical propensity of these transmembrane peptides that match perfectly their
experimental secondary structures, and we identified common motifs that reflect
their quaternary organization and known (or not) biochemical function. We
showed that BM2 is organized into two structurally distinct parts: a
significantly more stable N-terminal half, and a fast-converting C-terminal
half that continuously folds and unfolds between α-helical structures and
non-canonical structures, which are mostly turns. In AM2, both the N-terminal
half and C-terminal half are very flexible. In contrast, the two
coronavirus-derived transmembrane peptides are much more stable and fast
helix-formers when compared with the influenza ones. In particular, the
SARS-derived peptide E appears to be the fastest and most stable helix-former
of all the four viral peptides studied, with a helical structure that persists
almost without disruption for the whole of its 10 μs simulation. By comparing
the results with experimental observations, we benchmarked TFE in studying the
conformation of membrane and hydrophobic peptides. This work provided accurate
results suggesting a methodology to run long MD simulations and predict
structural properties of biologically important membrane peptides.
We have carried out a proof-of-concept molecular dynamics (MD) simulation
with adaptive tempering in a membrane mimetic environment to study the folding
of single-pass membrane peptides. We tested the influenza A M2 viroporin,
influenza B M2 viroporin, and protein E from coronaviruses MERS-Cov-2 and
SARS-CoV-2 peptides with known experimental secondary structures in membrane
bilayers. The two influenza-derived peptides are significantly different in the
peptide sequence and secondary structure and more polar than the two
coronavirus-derived peptides. Through a total of more than 50 μs of simulation
time that could be accomplished in trifluoroethanol (TFE), as a membrane model,
we characterized comparatively the folding behavior, helical stability, and
helical propensity of these transmembrane peptides that match perfectly their
experimental secondary structures, and we identified common motifs that reflect
their quaternary organization and known (or not) biochemical function. We
showed that BM2 is organized into two structurally distinct parts: a
significantly more stable N-terminal half, and a fast-converting C-terminal
half that continuously folds and unfolds between α-helical structures and
non-canonical structures, which are mostly turns. In AM2, both the N-terminal
half and C-terminal half are very flexible. In contrast, the two
coronavirus-derived transmembrane peptides are much more stable and fast
helix-formers when compared with the influenza ones. In particular, the
SARS-derived peptide E appears to be the fastest and most stable helix-former
of all the four viral peptides studied, with a helical structure that persists
almost without disruption for the whole of its 10 μs simulation. By comparing
the results with experimental observations, we benchmarked TFE in studying the
conformation of membrane and hydrophobic peptides. This work provided accurate
results suggesting a methodology to run long MD simulations and predict
structural properties of biologically important membrane peptides.
52. Gkogka, I. & Glykos*, N.M. (2022), "Folding molecular dynamics simulation of T-peptide, a HIV viral entry inhibitor : Structure, dynamics, and comparison with the experimental data", J. Comput. Chem., 43, 942-952.
Electronic reprint (18 MBytes) : Local copy, or directly from J. Comput. Chem., Copyright © Wiley Periodicals, Inc.
 Peptide T is a synthetic octapeptide fragment, which corresponds to the region
185–192 of the gp120 HIV coat protein and functions as a viral entry inhibitor.
In this work, a folding molecular dynamics simulation of peptide T in a
membrane-mimicking (DMSO) solution was performed with the aim of characterizing
the peptide's structural and dynamical properties. We show that peptide T is
highly flexible and dynamic. The main structural characteristics observed were
rapidly interconverting short helical stretches and turns, with a notable
preference for the formation of β-turns. The simulation also indicated that the
C-terminal part appears to be more stable than the rest of the peptide, with the
most preferred conformation for residues 5–8 being a β-turn. In order to
validate the accuracy of the simulations, we compared our results with the
experimental NMR data obtained for the T-peptide in the same solvent. In
agreement with the simulation, the NMR data indicated the presence of a
preferred structure in solution that was consistent with a β-turn comprising the
four C-terminal residues. An additional comparison between the experimental and
simulation-derived chemical shifts also showed a reasonable agreement between
experiment and simulation, further validating the simulation-derived structural
characterization of the T-peptide. We conclude that peptide folding simulations
produce physically relevant results even when performed with organic solvents
that were not part of the force field parameterization procedure.
Peptide T is a synthetic octapeptide fragment, which corresponds to the region
185–192 of the gp120 HIV coat protein and functions as a viral entry inhibitor.
In this work, a folding molecular dynamics simulation of peptide T in a
membrane-mimicking (DMSO) solution was performed with the aim of characterizing
the peptide's structural and dynamical properties. We show that peptide T is
highly flexible and dynamic. The main structural characteristics observed were
rapidly interconverting short helical stretches and turns, with a notable
preference for the formation of β-turns. The simulation also indicated that the
C-terminal part appears to be more stable than the rest of the peptide, with the
most preferred conformation for residues 5–8 being a β-turn. In order to
validate the accuracy of the simulations, we compared our results with the
experimental NMR data obtained for the T-peptide in the same solvent. In
agreement with the simulation, the NMR data indicated the presence of a
preferred structure in solution that was consistent with a β-turn comprising the
four C-terminal residues. An additional comparison between the experimental and
simulation-derived chemical shifts also showed a reasonable agreement between
experiment and simulation, further validating the simulation-derived structural
characterization of the T-peptide. We conclude that peptide folding simulations
produce physically relevant results even when performed with organic solvents
that were not part of the force field parameterization procedure.
51. Mitsikas, D.A. & Glykos*, N.M. (2020), "A molecular dynamics simulation study on the propensity of Asn-Gly-containing heptapeptides towards β-turn structures: Comparison with ab initio quantum mechanical calculations", PLoS ONE, 15(12): e0243429.
Electronic reprint (2.8 MBytes) : Local copy, or directly from PLoS ONE, Copyright © Mitsikas & Glykos under the Creative Commons Attribution License.

 Both molecular mechanical and quantum mechanical calculations play an important
role in describing the behavior and structure of molecules. In this work, we
compare for the same peptide systems the results obtained from folding molecular
dynamics simulations with previously reported results from quantum mechanical
calculations. More specifically, three molecular dynamics simulations of 5 μs
each in explicit water solvent were carried out for three Asn-Gly-containing
heptapeptides, in order to study their folding and dynamics. Previous data,
based on quantum mechanical calculations within the DFT framework have shown
that these peptides adopt β-turn structures in aqueous solution, with type I’
β-turn being the most preferred motif. The results from our analyses indicate
that at least for the given systems, force field and simulation protocol, the
two methods diverge in their predictions. The possibility of a force
field-dependent deficiency is examined as a possible source of the observed
discrepancy.
Both molecular mechanical and quantum mechanical calculations play an important
role in describing the behavior and structure of molecules. In this work, we
compare for the same peptide systems the results obtained from folding molecular
dynamics simulations with previously reported results from quantum mechanical
calculations. More specifically, three molecular dynamics simulations of 5 μs
each in explicit water solvent were carried out for three Asn-Gly-containing
heptapeptides, in order to study their folding and dynamics. Previous data,
based on quantum mechanical calculations within the DFT framework have shown
that these peptides adopt β-turn structures in aqueous solution, with type I’
β-turn being the most preferred motif. The results from our analyses indicate
that at least for the given systems, force field and simulation protocol, the
two methods diverge in their predictions. The possibility of a force
field-dependent deficiency is examined as a possible source of the observed
discrepancy.
50. Kokkinidis, M., Glykos*, N.M. & Fadouloglou*, V.E. (2020), "Catalytic activity regulation through post-translational modification: the expanding universe of protein diversity", Adv. Protein Chem. Struct. Biol., 122, 97-125.
Electronic reprint (2.5 MBytes) : Local copy, or directly from Adv. Protein Chem. Struct. Biol., Copyright © Elsevier.
 Protein composition is restricted by the genetic code to a relatively small
number of natural amino acids. Similarly, the known three-dimensional structures
adopt a limited number of protein folds. However, proteins exert a large variety
of functions and show a remarkable ability for regulation and immediate response
to intracellular and extracellular stimuli. To some degree, the wide variability
of protein function can be attributed to the post-translational modifications.
Post-translational modifications have been observed in all kingdoms of life and
give to proteins a significant degree of chemical and consequently functional
and structural diversity. Their importance is partly reflected in the large
number of genes dedicated to their regulation. So far, hundreds of
post-translational modifications have been observed while it is believed that
many more are to be discovered along with the technological advances in
sequencing, proteomics, mass spectrometry and structural biology. Indeed, the
number of studies which report novel post translational modifications is getting
larger supporting the notion that their space is still largely unexplored. In
this review we explore the impact of post-translational modifications on protein
structure and function with emphasis on catalytic activity regulation. We
present examples of proteins and protein families whose catalytic activity is
substantially affected by the presence of post translational modifications and
we describe the molecular basis which underlies the regulation of the protein
function through these modifications. When available, we also summarize the
current state of knowledge on the mechanisms which introduce these modifications
to protein sites.
Protein composition is restricted by the genetic code to a relatively small
number of natural amino acids. Similarly, the known three-dimensional structures
adopt a limited number of protein folds. However, proteins exert a large variety
of functions and show a remarkable ability for regulation and immediate response
to intracellular and extracellular stimuli. To some degree, the wide variability
of protein function can be attributed to the post-translational modifications.
Post-translational modifications have been observed in all kingdoms of life and
give to proteins a significant degree of chemical and consequently functional
and structural diversity. Their importance is partly reflected in the large
number of genes dedicated to their regulation. So far, hundreds of
post-translational modifications have been observed while it is believed that
many more are to be discovered along with the technological advances in
sequencing, proteomics, mass spectrometry and structural biology. Indeed, the
number of studies which report novel post translational modifications is getting
larger supporting the notion that their space is still largely unexplored. In
this review we explore the impact of post-translational modifications on protein
structure and function with emphasis on catalytic activity regulation. We
present examples of proteins and protein families whose catalytic activity is
substantially affected by the presence of post translational modifications and
we describe the molecular basis which underlies the regulation of the protein
function through these modifications. When available, we also summarize the
current state of knowledge on the mechanisms which introduce these modifications
to protein sites.
49. Stylianakis, I., Shalev, A., Scheiner, S., Sigalas, M.P., Arkin, I.T., Glykos*, N.M. & Kolocouris*, A. (2020), "The balance between side‐chain and backbone‐driven association in folding of the α‐helical influenza A transmembrane peptide", J. Comput. Chem., 41, 2177-2188.
Electronic reprint (3.1 MBytes) : Local copy, or directly from J. Comput. Chem., Copyright © Wiley Periodicals, Inc.

 The correct balance between attractive, repulsive and peptide hydrogen bonding
interactions must be attained for proteins to fold correctly. To investigate
these important contributors, we sought a comparison of the folding between two
25‐residues peptides, the influenza A M2 protein transmembrane domain (M2TM) and
the 25‐Ala (Ala25). M2TM forms a stable α‐helix as is shown by circular
dichroism (CD) experiments. Molecular dynamics (MD) simulations with adaptive
tempering show that M2TM monomer is more dynamic in nature and quickly
interconverts between an ensemble of various α‐helical structures, and less
frequently turns and coils, compared to one α‐helix for Ala25. DFT calculations
suggest that folding from the extended structure to the α‐helical structure is
favored for M2TM compared with Ala25. This is due to CH⋯O attractive
interactions which favor folding to the M2TM α‐helix, and cannot be described
accurately with a force field. Using natural bond orbital (NBO) analysis and
quantum theory atoms in molecules (QTAIM) calculations, 26 CH⋯O interactions and
22 NH⋯O hydrogen bonds are calculated for M2TM. The calculations show that CH⋯O
hydrogen bonds, although individually weaker, have a cumulative effect that
cannot be ignored and may contribute as much as half of the total hydrogen
bonding energy, when compared to NH⋯O, to the stabilization of the α‐helix in
M2TM. Further, a strengthening of NH⋯O hydrogen bonding interactions is
calculated for M2TM compared to Ala25. Additionally, these weak CH⋯O
interactions can dissociate and associate easily leading to the ensemble of
folded structures for M2TM observed in folding MD simulations.
The correct balance between attractive, repulsive and peptide hydrogen bonding
interactions must be attained for proteins to fold correctly. To investigate
these important contributors, we sought a comparison of the folding between two
25‐residues peptides, the influenza A M2 protein transmembrane domain (M2TM) and
the 25‐Ala (Ala25). M2TM forms a stable α‐helix as is shown by circular
dichroism (CD) experiments. Molecular dynamics (MD) simulations with adaptive
tempering show that M2TM monomer is more dynamic in nature and quickly
interconverts between an ensemble of various α‐helical structures, and less
frequently turns and coils, compared to one α‐helix for Ala25. DFT calculations
suggest that folding from the extended structure to the α‐helical structure is
favored for M2TM compared with Ala25. This is due to CH⋯O attractive
interactions which favor folding to the M2TM α‐helix, and cannot be described
accurately with a force field. Using natural bond orbital (NBO) analysis and
quantum theory atoms in molecules (QTAIM) calculations, 26 CH⋯O interactions and
22 NH⋯O hydrogen bonds are calculated for M2TM. The calculations show that CH⋯O
hydrogen bonds, although individually weaker, have a cumulative effect that
cannot be ignored and may contribute as much as half of the total hydrogen
bonding energy, when compared to NH⋯O, to the stabilization of the α‐helix in
M2TM. Further, a strengthening of NH⋯O hydrogen bonding interactions is
calculated for M2TM compared to Ala25. Additionally, these weak CH⋯O
interactions can dissociate and associate easily leading to the ensemble of
folded structures for M2TM observed in folding MD simulations.
48. Riziotis, I.G. & Glykos*, N.M. (2019), "On the presence of short-range periodicities in protein structures that are not related to established secondary structure elements", Proteins, 87, 966-978.
Electronic reprint (5.7 MBytes) : Local copy, or directly from Proteins, Copyright © Wiley. See also the Supporting information file (2.3 Mbytes).

 Standard secondary structure elements such as α-helices or β-sheets, are characterized by repeating
backbone torsion angles (φ,ψ) at the single residue level. Two-residue motifs of the type (φ,ψ)₂ are
also observed in non-linear conformations, mainly turns. Taking these observations a step further, it
can be argued that there is no a priori reason why the presence of higher order periodicities can not
be envisioned in protein structures, such as, for example, periodic transitions between successive
residues of the type (...-α-β-α-β-α-...), or, (...-β-αL-β-αL-β-...), or, (...-α-β-αL-α-β-αL-...), etc., where
the symbols (α,β,αL) refer to the established Ramachandran-based residue conformations. From all
such possible higher order periodicities, here we examine the deposited (with the PDB) protein
structures for the presence of short-range periodical conformations comprising five consecutive
residues alternating between two (and only two) distinct Ramachandran regions, for example
conformations of the type (α-β-α-β-α) or (β-αL-β-αL-β) etc. Using a probabilistic approach we have
located several thousand of such peptapeptides, and these were clustered and analyzed in terms of
their structural characteristics, their sequences, and their putative functional correlations using a
gene ontology-based approach. We show that such non-standard short-range periodicities are
present in a large and functionally diverse sample of proteins, and can be grouped into two
structurally conserved major types. Examination of the structural context in which these
peptapeptides are observed gave no conclusive evidence for the presence of a persistent structural or
functional role of these higher order periodic conformations.
Standard secondary structure elements such as α-helices or β-sheets, are characterized by repeating
backbone torsion angles (φ,ψ) at the single residue level. Two-residue motifs of the type (φ,ψ)₂ are
also observed in non-linear conformations, mainly turns. Taking these observations a step further, it
can be argued that there is no a priori reason why the presence of higher order periodicities can not
be envisioned in protein structures, such as, for example, periodic transitions between successive
residues of the type (...-α-β-α-β-α-...), or, (...-β-αL-β-αL-β-...), or, (...-α-β-αL-α-β-αL-...), etc., where
the symbols (α,β,αL) refer to the established Ramachandran-based residue conformations. From all
such possible higher order periodicities, here we examine the deposited (with the PDB) protein
structures for the presence of short-range periodical conformations comprising five consecutive
residues alternating between two (and only two) distinct Ramachandran regions, for example
conformations of the type (α-β-α-β-α) or (β-αL-β-αL-β) etc. Using a probabilistic approach we have
located several thousand of such peptapeptides, and these were clustered and analyzed in terms of
their structural characteristics, their sequences, and their putative functional correlations using a
gene ontology-based approach. We show that such non-standard short-range periodicities are
present in a large and functionally diverse sample of proteins, and can be grouped into two
structurally conserved major types. Examination of the structural context in which these
peptapeptides are observed gave no conclusive evidence for the presence of a persistent structural or
functional role of these higher order periodic conformations.
47. Georgoulia, P.S. & Glykos*, N.M. (2019), "Molecular simulation of peptides coming of age: Accurate prediction of folding, dynamics and structures", Arch. Biochem. Biophys., 664, 76-88.
Electronic reprint (1.6 MBytes) : Local copy, or directly from Arch. Biochem. Biophys., Copyright © Elsevier Inc.

 The application of molecular dynamics simulations to study the folding and
dynamics of peptides has attracted a lot of interest in the last couple of
decades. Following the successful prediction of the folding of several proteins
using molecular simulation, foldable peptides emerged as a favourable system
mainly due to their application in improving protein structure prediction
methods and in drug design studies. However, their performance is inherently
linked to the accuracy of the empirical force fields used in the simulations,
whose optimisation and validation is of paramount importance. Here we review
the most important findings in the field of molecular peptide simulations and
highlight the significant advancements made over the last twenty years. Special
reference is made on the simulation of disordered peptides and the remaining
challenge to find a force field able to describe accurately their
conformational landscape.
The application of molecular dynamics simulations to study the folding and
dynamics of peptides has attracted a lot of interest in the last couple of
decades. Following the successful prediction of the folding of several proteins
using molecular simulation, foldable peptides emerged as a favourable system
mainly due to their application in improving protein structure prediction
methods and in drug design studies. However, their performance is inherently
linked to the accuracy of the empirical force fields used in the simulations,
whose optimisation and validation is of paramount importance. Here we review
the most important findings in the field of molecular peptide simulations and
highlight the significant advancements made over the last twenty years. Special
reference is made on the simulation of disordered peptides and the remaining
challenge to find a force field able to describe accurately their
conformational landscape.
46. Georgoulia*, P.S. & Glykos, N.M. (2018), "Folding Molecular Dynamics Simulation of a gp41-Derived Peptide Reconcile Divergent Structure Determinations", ACS Omega, 3, 14746-14754.
Electronic reprint (1.7 MBytes) : Local copy, or directly from ACS Omega, Copyright © American Chemical Society. See also the Supporting information file (11 Mbytes).

 T-20 peptide is the first FDA-approved fusion inhibitor against AIDS/HIV-1 gp41
protein. Part of it, the gp41[659-671] peptide, that contains the complete
epitope for the neutralizing 2F5 monoclonal antibody, has been found
experimentally in a number of divergent structures. Herein, we attempt to
reconcile them by using unbiased large-scale all-atom molecular dynamics
folding simulations. We show that our approach can successfully capture the
peptide's heterogeneity and reach each and every experimentally determined
conformation in sub-angstrom accuracy, whilst preserving the peptide's
disordered nature. Our analysis also unveils that the minor refinements within
the AMBER99SB family of force fields can lead to appreciable differences in the
predicted conformational stability arising from subtle differences in the
helical- and β-region of the Ramachandran plot. Our work underlines the
contribution of molecular dynamics simulation in structurally characterizing
pharmacologically important peptides of ambiguous structure.
T-20 peptide is the first FDA-approved fusion inhibitor against AIDS/HIV-1 gp41
protein. Part of it, the gp41[659-671] peptide, that contains the complete
epitope for the neutralizing 2F5 monoclonal antibody, has been found
experimentally in a number of divergent structures. Herein, we attempt to
reconcile them by using unbiased large-scale all-atom molecular dynamics
folding simulations. We show that our approach can successfully capture the
peptide's heterogeneity and reach each and every experimentally determined
conformation in sub-angstrom accuracy, whilst preserving the peptide's
disordered nature. Our analysis also unveils that the minor refinements within
the AMBER99SB family of force fields can lead to appreciable differences in the
predicted conformational stability arising from subtle differences in the
helical- and β-region of the Ramachandran plot. Our work underlines the
contribution of molecular dynamics simulation in structurally characterizing
pharmacologically important peptides of ambiguous structure.
45. Adamidou, T., Arvaniti, K.-O. & Glykos*, N.M. (2018), "Folding Simulations of a Nuclear Receptor Box-Containing Peptide Demonstrate the Structural Persistence of the LxxLL Motif Even in the Absence of Its Cognate Receptor", J. Phys. Chem. B, 122, 106â116.
Electronic reprint (2.5 MBytes) : Local copy, or directly from J. Phys. Chem. B, Copyright © American Chemical Society. See also the Supporting information file (3.3 Mbytes).
 Regulation of nuclear receptors by their coactivators involves the
recognition and binding of a specific sequence motif contained in the
coactivator sequence. This motif is known as the nuclear receptor (NR) box
and contains a conserved LxxLL subsequence, where L is leucine and x is any
amino acid residue. Crystallographic studies have shown that the LxxLL
motifs adopt an α-helical conformation when bound to their cognate nuclear
receptors. Here we use an extensive set of folding molecular dynamics
simulations to examine whether the α-helical conformation demonstrated by
the LxxLL motifs in the bound state may represent a persistent structural
preference of these peptides even in the absence of their cognate receptors.
To this end, we have performed a grand total of 35 ÎŒs of adaptive tempering
folding simulations of an NR-box-containing peptide derived from
Drosophila's fushi tarazu segmentation gene product. Our
simulationsâperformed using full electrostatics and an explicit
representation of two different solvents (water and a TFE/water
mixture)âclearly indicate the presence of a persistent helical preference of
the LxxLL motif with a concomitant native-like structure and contacts
between the motif's leucine residues. To lend further support to our
findings, we compare the simulation-derived peptide dynamics with
experimental NMR-derived nuclear Overhauser effect (NOE) measurements that
had been previously obtained for the same peptide in the same two solvents.
The comparison demonstrates a quantitative agreement between simulation and
experiment with average upper bound NOE violations of less than 0.084 Ã
,
thus independently validating our main conclusion concerning the intrinsic
preference of NR-box motifs to form helical structures even in the absence
of their cognate receptors.
Regulation of nuclear receptors by their coactivators involves the
recognition and binding of a specific sequence motif contained in the
coactivator sequence. This motif is known as the nuclear receptor (NR) box
and contains a conserved LxxLL subsequence, where L is leucine and x is any
amino acid residue. Crystallographic studies have shown that the LxxLL
motifs adopt an α-helical conformation when bound to their cognate nuclear
receptors. Here we use an extensive set of folding molecular dynamics
simulations to examine whether the α-helical conformation demonstrated by
the LxxLL motifs in the bound state may represent a persistent structural
preference of these peptides even in the absence of their cognate receptors.
To this end, we have performed a grand total of 35 ÎŒs of adaptive tempering
folding simulations of an NR-box-containing peptide derived from
Drosophila's fushi tarazu segmentation gene product. Our
simulationsâperformed using full electrostatics and an explicit
representation of two different solvents (water and a TFE/water
mixture)âclearly indicate the presence of a persistent helical preference of
the LxxLL motif with a concomitant native-like structure and contacts
between the motif's leucine residues. To lend further support to our
findings, we compare the simulation-derived peptide dynamics with
experimental NMR-derived nuclear Overhauser effect (NOE) measurements that
had been previously obtained for the same peptide in the same two solvents.
The comparison demonstrates a quantitative agreement between simulation and
experiment with average upper bound NOE violations of less than 0.084 Ã
,
thus independently validating our main conclusion concerning the intrinsic
preference of NR-box motifs to form helical structures even in the absence
of their cognate receptors.
44. Fadouloglou, V.E., Balomenou, S., Aivaliotis, M., Kotsifaki, D., Arnaouteli, S., Tomatsidou, A., Efstathiou, G., Kountourakis, N., Miliara, S., Griniezaki, M., Tsalafouta, A., Pergantis, S.A., Boneca, I.G., Glykos, N.M., Bouriotis, V. & Kokkinidis*, M. (2017), "An unusual α-carbon hydroxylation of proline promotes active-site maturation", J. Am. Chem. Soc., 139, 5330â5337.
Electronic reprint (2.5 MBytes) : Local copy, or directly from J. Am. Chem. Soc., Copyright © American Chemical Society.
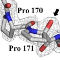
 The full extent of proline (Pro) hydroxylation has yet to be established, as it is largely
unexplored in bacteria. We describe here a so far unknown Pro hydroxylation activity which occurs in
active sites of polysaccharide deacetylases (PDAs) from bacterial pathogens, modifying the protein
backbone at the Cα atom of a Pro residue to produce 2-hydroxyproline (2-Hyp). This process modifies with
high specificity a conserved Pro, shares with the deacetylation reaction the same active site and one catalytic
residue and utilizes molecular oxygen as source for the hydroxyl group oxygen of 2-Hyp. By
providing additional hydrogen bonding capacity, the Pro â 2-Hyp conversion alters the active site and
enhances significantly deacetylase activity, probably by creating a more favorable environment for
transition state stabilization. Our results classify this process as an active site "maturation", which is
highly atypical by being a protein backbone modifying activity, rather than a side-chain modifying one.
The full extent of proline (Pro) hydroxylation has yet to be established, as it is largely
unexplored in bacteria. We describe here a so far unknown Pro hydroxylation activity which occurs in
active sites of polysaccharide deacetylases (PDAs) from bacterial pathogens, modifying the protein
backbone at the Cα atom of a Pro residue to produce 2-hydroxyproline (2-Hyp). This process modifies with
high specificity a conserved Pro, shares with the deacetylation reaction the same active site and one catalytic
residue and utilizes molecular oxygen as source for the hydroxyl group oxygen of 2-Hyp. By
providing additional hydrogen bonding capacity, the Pro â 2-Hyp conversion alters the active site and
enhances significantly deacetylase activity, probably by creating a more favorable environment for
transition state stabilization. Our results classify this process as an active site "maturation", which is
highly atypical by being a protein backbone modifying activity, rather than a side-chain modifying one.
43. Serafeim A.-P., Salamanos, G., Patapati, K.K. & Glykos*, N.M. (2016), "Sensitivity of Folding Molecular Dynamics Simulations to Even Minor Force Field Changes", J. Chem. Inf. Model., 56, 2035-2041.
Electronic reprint (2.6 MBytes) : Local copy, or directly from J. Chem. Inf. Model., Copyright © American Chemical Society. See also the Supporting information file.

 We examine the sensitivity of folding molecular dynamics simulations on the
choice between three variants of the same force field (the AMBER99SB force
field and its ILDN, NMR-ILDN and STAR-ILDN variants). Using two different
peptide systems (a marginally stable helical peptide and a β-hairpin) and a
grand total of more than 20 ÎŒs of simulation time we show that even
relatively minor force field changes can lead to appreciable differences in
the peptide folding behavior.
We examine the sensitivity of folding molecular dynamics simulations on the
choice between three variants of the same force field (the AMBER99SB force
field and its ILDN, NMR-ILDN and STAR-ILDN variants). Using two different
peptide systems (a marginally stable helical peptide and a β-hairpin) and a
grand total of more than 20 ÎŒs of simulation time we show that even
relatively minor force field changes can lead to appreciable differences in
the peptide folding behavior.
42. Baltzis, A.S. & Glykos*, N.M. (2016), "Characterizing a partially ordered miniprotein through folding molecular dynamics simulations: Comparison with the experimental data", Prot. Sci., 25, 587â596.
Electronic reprint (3.3 MBytes) : Local copy, or directly from Protein Science, Copyright © Wiley Periodicals, Inc.

 The villin headpiece helical subdomain (HP36) is one of the best known model
systems for computational studies of fast-folding all-α miniproteins. HP21
is a peptide fragment âderived from HP36â comprising only the first and
second helices of the full domain. Experimental studies showed that although
HP21 is mostly unfolded in solution, it does maintain some persistent
native-like structure as indicated by the analysis of NMR-derived chemical
shifts. Here we compare the experimental data for HP21 with the results
obtained from a 15 ÎŒs long folding molecular dynamics simulation performed
in explicit water and with full electrostatics. We find that the simulation
is in good agreement with the experiment and faithfully reproduces the major
experimental findings, namely that (a) HP21 is disordered in solution with
less that 10% of the trajectory corresponding to transiently stable
structures, (b) the most highly populated conformer is a native-like
structure with an RMSD from the corresponding portion of the HP36 crystal
structure of less than 1Ã
, (c) the simulation-derived chemical shifts âover
the whole length of the trajectoryâ are in reasonable agreement with the
experiment giving reduced Ï2 values of 1.6, 1.4 and 0.8 for the ÎÎŽ13Cα,
ÎÎŽ13CO and ÎÎŽ13Cβ secondary shifts respectively (becoming 0.8, 0.7, and 0.3
when only the major peptide conformer is considered), and finally, (d) the
secondary structure propensity scores are in very good agreement with the
experiment and clearly indicate the higher stability of the first helix. We
conclude that folding molecular dynamics simulations can be a useful tool
for the structural characterization of even marginally stable peptides.
The villin headpiece helical subdomain (HP36) is one of the best known model
systems for computational studies of fast-folding all-α miniproteins. HP21
is a peptide fragment âderived from HP36â comprising only the first and
second helices of the full domain. Experimental studies showed that although
HP21 is mostly unfolded in solution, it does maintain some persistent
native-like structure as indicated by the analysis of NMR-derived chemical
shifts. Here we compare the experimental data for HP21 with the results
obtained from a 15 ÎŒs long folding molecular dynamics simulation performed
in explicit water and with full electrostatics. We find that the simulation
is in good agreement with the experiment and faithfully reproduces the major
experimental findings, namely that (a) HP21 is disordered in solution with
less that 10% of the trajectory corresponding to transiently stable
structures, (b) the most highly populated conformer is a native-like
structure with an RMSD from the corresponding portion of the HP36 crystal
structure of less than 1Ã
, (c) the simulation-derived chemical shifts âover
the whole length of the trajectoryâ are in reasonable agreement with the
experiment giving reduced Ï2 values of 1.6, 1.4 and 0.8 for the ÎÎŽ13Cα,
ÎÎŽ13CO and ÎÎŽ13Cβ secondary shifts respectively (becoming 0.8, 0.7, and 0.3
when only the major peptide conformer is considered), and finally, (d) the
secondary structure propensity scores are in very good agreement with the
experiment and clearly indicate the higher stability of the first helix. We
conclude that folding molecular dynamics simulations can be a useful tool
for the structural characterization of even marginally stable peptides.
41. Baltzis, A.S., Koukos, P.I. & Glykos*, N.M. (2015), "Clustering of molecular dynamics trajectories via peak-picking in multidimensional PCA-derived distributions", arXiv:1512.04024 [q-bio.BM].
Electronic reprint (826 KBytes) : Local copy, or directly from arXiv:1512.04024.

 We describe a robust, fast, and memory-efficient procedure that can cluster
millions of structures derived from molecular dynamics simulations. The
essence of the method is based on a peak-picking algorithm applied to three-
and five-dimensional distributions of the principal components derived from
the trajectory and automatically supports both Cartesian and dihedral
PCA-based clustering. The density threshold required for identifying
isolated peaks (which correspond to discrete clusters) is determined through
the application of a variance-explained criterion which allows for a
completely automated clustering procedure with no user intervention. In this
communication we describe the algorithm and present some of the results
obtained from the application of the method as implemented in the molecular
dynamics analysis programs carma, grcarma. and cluster5D. We conclude with a
discussion of the limitations and possible pitfalls of this method.
We describe a robust, fast, and memory-efficient procedure that can cluster
millions of structures derived from molecular dynamics simulations. The
essence of the method is based on a peak-picking algorithm applied to three-
and five-dimensional distributions of the principal components derived from
the trajectory and automatically supports both Cartesian and dihedral
PCA-based clustering. The density threshold required for identifying
isolated peaks (which correspond to discrete clusters) is determined through
the application of a variance-explained criterion which allows for a
completely automated clustering procedure with no user intervention. In this
communication we describe the algorithm and present some of the results
obtained from the application of the method as implemented in the molecular
dynamics analysis programs carma, grcarma. and cluster5D. We conclude with a
discussion of the limitations and possible pitfalls of this method.
40. Koukos, P.I. & Glykos*, N.M. (2014), "Folding Molecular Dynamics Simulations Accurately Predict the Effect of Mutations on the Stability and Structure of a Vammin-Derived Peptide", J. Phys. Chem. B, 118, 10076â10084.
Electronic reprint (3.7 MBytes) : Local copy, or directly from J. Phys. Chem. B, Copyright © American Chemical Society. See also the Supporting Information.

 Folding molecular dynamics simulations amounting to a grand total of 4 ÎŒs of
simulation time were performed on two peptides (with native and mutated
sequences) derived from loop 3 of the vammin protein and the results
compared with the experimentally known peptide stabilities and structures.
The simulations faithfully and accurately reproduce the major experimental
findings and show that (a) the native peptide is mostly disordered in
solution, (b) the mutant peptide has a well-defined and stable structure,
and (c) the structure of the mutant is an irregular β-hairpin with a
non-glycine β-bulge, in excellent agreement with the peptideâs known NMR
structure. Additionally, the simulations also predict the presence of a very
small β-hairpin-like population for the native peptide but surprisingly
indicate that this population is structurally more similar to the structure
of the native peptide as observed in the vammin protein than to the NMR
structure of the isolated mutant peptide. We conclude that, at least for the
given system, force field, and simulation protocol, folding molecular
dynamics simulations appear to be successful in reproducing the
experimentally accessible physical reality to a satisfactory level of detail
and accuracy.
Folding molecular dynamics simulations amounting to a grand total of 4 ÎŒs of
simulation time were performed on two peptides (with native and mutated
sequences) derived from loop 3 of the vammin protein and the results
compared with the experimentally known peptide stabilities and structures.
The simulations faithfully and accurately reproduce the major experimental
findings and show that (a) the native peptide is mostly disordered in
solution, (b) the mutant peptide has a well-defined and stable structure,
and (c) the structure of the mutant is an irregular β-hairpin with a
non-glycine β-bulge, in excellent agreement with the peptideâs known NMR
structure. Additionally, the simulations also predict the presence of a very
small β-hairpin-like population for the native peptide but surprisingly
indicate that this population is structurally more similar to the structure
of the native peptide as observed in the vammin protein than to the NMR
structure of the isolated mutant peptide. We conclude that, at least for the
given system, force field, and simulation protocol, folding molecular
dynamics simulations appear to be successful in reproducing the
experimentally accessible physical reality to a satisfactory level of detail
and accuracy.
39. Koukos, P.I. & Glykos*, N.M. (2014), "On the application of Good-Turing statistics to quantify convergence of biomolecular simulations", J. Chem. Inf. Model., 54, 209-217.
Electronic reprint (1.1 MBytes) : Local copy, or directly from J. Chem. Inf. Model., Copyright © American Chemical Society.

 Quantifying convergence and sufficient sampling of macromolecular molecular
dynamics simulations is more often than not a source of controversy (and of
various ad hoc solutions) in the field. Clearly, the only reasonable,
consistent and satisfying way to infer convergence (or otherwise) of a
molecular dynamics trajectory must be based on probability theory. Ideally,
the question we would wish to answer is the following : "What is the
probability that a molecular configuration important for the analysis in
hand has not yet been observed ?". Here we propose a method for answering a
variant of this question by using the Good-Turing formalism for frequency
estimation of unobserved species in a sample. Although several approaches
may be followed in order to deal with the problem of discretizing the
configurational space, for this work we use the classical RMSD matrix as a
means to answering the following question : "What is the probability that a
molecular configuration with an RMSD (from all other already observed
configurations) higher than a given threshold has not actually been observed
?". We apply the proposed method to several different trajectories and show
that the procedure appears to be both computationally stable and internally
consistent. A free, open-source program implementing these ideas is
immediately available for download via public repositories.
Quantifying convergence and sufficient sampling of macromolecular molecular
dynamics simulations is more often than not a source of controversy (and of
various ad hoc solutions) in the field. Clearly, the only reasonable,
consistent and satisfying way to infer convergence (or otherwise) of a
molecular dynamics trajectory must be based on probability theory. Ideally,
the question we would wish to answer is the following : "What is the
probability that a molecular configuration important for the analysis in
hand has not yet been observed ?". Here we propose a method for answering a
variant of this question by using the Good-Turing formalism for frequency
estimation of unobserved species in a sample. Although several approaches
may be followed in order to deal with the problem of discretizing the
configurational space, for this work we use the classical RMSD matrix as a
means to answering the following question : "What is the probability that a
molecular configuration with an RMSD (from all other already observed
configurations) higher than a given threshold has not actually been observed
?". We apply the proposed method to several different trajectories and show
that the procedure appears to be both computationally stable and internally
consistent. A free, open-source program implementing these ideas is
immediately available for download via public repositories.
38. Koukos, P.I. & Glykos*, N.M. (2013), "grcarma: A Fully Automated Task-Oriented Interface for the Analysis of Molecular Dynamics Trajectories", J. Comput. Chem., 34, 2310-2312, Cover story for Vol.34, Issue 26.
Electronic reprint (929 KBytes) : Local copy, or directly from J. Comput. Chem., Copyright © Wiley Periodicals, Inc.

 We report the availability of grcarma, a program encoding for
a fully automated set of tasks aiming to simplify the analysis
of molecular dynamics trajectories of biological macromolecules.
It is a cross-platform, Perl/Tk-based front-end to the program
carma and is designed to facilitate the needs of the
novice as well as those of the expert user, while at the same
time maintaining a user-friendly and intuitive design. Particular
emphasis was given to the automation of several tedious
tasks, such as extraction of clusters of structures based on
dihedral and Cartesian principal component analysis, secondary
structure analysis, calculation and display of root-meansquare
deviation (RMSD) matrices, calculation of entropy, calculation
and analysis of varianceâcovariance matrices, calculation
of the fraction of native contacts, etc. The program is
free-open source software available immediately for download.
We report the availability of grcarma, a program encoding for
a fully automated set of tasks aiming to simplify the analysis
of molecular dynamics trajectories of biological macromolecules.
It is a cross-platform, Perl/Tk-based front-end to the program
carma and is designed to facilitate the needs of the
novice as well as those of the expert user, while at the same
time maintaining a user-friendly and intuitive design. Particular
emphasis was given to the automation of several tedious
tasks, such as extraction of clusters of structures based on
dihedral and Cartesian principal component analysis, secondary
structure analysis, calculation and display of root-meansquare
deviation (RMSD) matrices, calculation of entropy, calculation
and analysis of varianceâcovariance matrices, calculation
of the fraction of native contacts, etc. The program is
free-open source software available immediately for download.
37. Kontopoulos, D.-G. & Glykos*, N.M. (2013), "Pinda: A Web service for detection and analysis of intraspecies gene duplication events", Comput. Methods Programs Biomed., 111, 711-714.
Electronic reprint (729 KBytes) : Local copy, or directly from Comput. Methods Programs Biomed., Copyright © Elsevier Ltd.

 We present Pinda, a Web service for the detection and analysis of possible
duplications of a given protein or DNA sequence within a source species.
Pinda fully automates the whole gene duplication detection procedure, from
performing the initial similarity searches, to generating the multiple
sequence alignments and the corresponding phylogenetic trees,
to bootstrapping the trees and producing a Z-score-based list of duplication
candidates for the input sequence. Pinda has been cross-validated using an
extensive set of known and bibliographically characterized duplication
events. The service facilitates the automatic and dependable identification
of gene duplication events, using some of the most successful bioinformatics
software to perform an extensive analysis protocol. Pinda will prove of
use for the analysis of newly discovered genes and proteins, thus also
assisting the study of recently sequenced genomes. The service's location is
http://orion.mbg.duth.gr/Pinda. The source code is freely available via
https://github.com/dgkontopoulos/Pinda/.
We present Pinda, a Web service for the detection and analysis of possible
duplications of a given protein or DNA sequence within a source species.
Pinda fully automates the whole gene duplication detection procedure, from
performing the initial similarity searches, to generating the multiple
sequence alignments and the corresponding phylogenetic trees,
to bootstrapping the trees and producing a Z-score-based list of duplication
candidates for the input sequence. Pinda has been cross-validated using an
extensive set of known and bibliographically characterized duplication
events. The service facilitates the automatic and dependable identification
of gene duplication events, using some of the most successful bioinformatics
software to perform an extensive analysis protocol. Pinda will prove of
use for the analysis of newly discovered genes and proteins, thus also
assisting the study of recently sequenced genomes. The service's location is
http://orion.mbg.duth.gr/Pinda. The source code is freely available via
https://github.com/dgkontopoulos/Pinda/.
36. Georgoulia, P.S. & Glykos*, N.M. (2013), "On the Foldability of Tryptophan-Containing Tetra- and Pentapeptides: An Exhaustive Molecular Dynamics Study", J. Phys. Chem. B, 117, 5522â5532.
Electronic reprint (3.1 MBytes) : Local copy, or directly from J. Phys. Chem. B, Copyright © American Chemical Society. See also the Supporting Information.

 Short peptides serve as minimal model systems
to decipher the determinants of foldability due to their
simplicity arising from their smaller size, their ability to echo
protein-like structural characteristics, and their direct implication
in force field validation. Here, we describe an effort to
identify small peptides that can still form stable structures in
aqueous solutions. We followed the in silico folding of a
selected set of 8640 tryptophan-containing tetra- and
pentapeptides through 15 210 molecular dynamics simulations
amounting to a total of 272.46 ÎŒs using explicit representation
of the solute and full treatment of the electrostatics. The
evaluation and sorting of peptides is achieved through scoring
functions, which include terms based on interatomic vector distances, atomic
fluctuations, and rmsd matrices between successive
frames of a trajectory. Highly scored peptides are studied further via
successive simulation rounds of increasing simulation length
and using different empirical force fields. Our method suggested only a
handful of peptides with strong foldability prognosis. The
discrepancies between the predictions of the various force fields for such
short sequences are also extensively discussed. We
conclude that the vast majority of such short peptides do not adopt
significantly stable structures in water solutions, at least based
on our computational predictions. The present work can be utilized in the
rational design and engineering of bioactive peptides
with desired molecular properties.
Short peptides serve as minimal model systems
to decipher the determinants of foldability due to their
simplicity arising from their smaller size, their ability to echo
protein-like structural characteristics, and their direct implication
in force field validation. Here, we describe an effort to
identify small peptides that can still form stable structures in
aqueous solutions. We followed the in silico folding of a
selected set of 8640 tryptophan-containing tetra- and
pentapeptides through 15 210 molecular dynamics simulations
amounting to a total of 272.46 ÎŒs using explicit representation
of the solute and full treatment of the electrostatics. The
evaluation and sorting of peptides is achieved through scoring
functions, which include terms based on interatomic vector distances, atomic
fluctuations, and rmsd matrices between successive
frames of a trajectory. Highly scored peptides are studied further via
successive simulation rounds of increasing simulation length
and using different empirical force fields. Our method suggested only a
handful of peptides with strong foldability prognosis. The
discrepancies between the predictions of the various force fields for such
short sequences are also extensively discussed. We
conclude that the vast majority of such short peptides do not adopt
significantly stable structures in water solutions, at least based
on our computational predictions. The present work can be utilized in the
rational design and engineering of bioactive peptides
with desired molecular properties.
35. Patmanidis, I. & Glykos*, N.M. (2013), "As good as it gets? Folding molecular dynamics simulations of the LytA choline-binding peptide result to an exceptionally accurate model of the peptide structure", J. Mol. Graph. Model., 41, 68-71.
Electronic reprint (686 KBytes) : Local copy, or directly from J. Mol. Graph. Model., 41, 68-71, Copyright © Elsevier Inc.
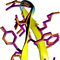
 Folding simulations of a choline-binding peptide derived from the Streptococcus pneumoniae LytA protein
converged to a model of the peptide's folded state structure which is in outstanding agreement with the
experimentally-determined structures, reaching values for the root mean squared deviation as low as
0.24 Ã
for the peptide's backbone atoms and 0.65 Ã
for all non-hydrogen atoms.
Folding simulations of a choline-binding peptide derived from the Streptococcus pneumoniae LytA protein
converged to a model of the peptide's folded state structure which is in outstanding agreement with the
experimentally-determined structures, reaching values for the root mean squared deviation as low as
0.24 Ã
for the peptide's backbone atoms and 0.65 Ã
for all non-hydrogen atoms.
34. Fadouloglou, V.E., Kapanidou, M., Agiomirgianaki, A., Arnaouteli, S., Bouriotis, V., Glykos*, N.M. & Kokkinidis*, M. (2013), "Structure determination through homology modelling and torsion-angle simulated annealing: application to a polysaccharide deacetylase from Bacillus cereus", Acta Crystallogr., D69, 276-283.
Electronic reprint (2.4 MBytes) : Local copy, or directly from Acta Crystallogr., D69, 276-283, Copyright © International Union of Crystallography.
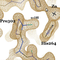
 The structure of BC0361, a polysaccharide deacetylase from
Bacillus cereus, has been determined using an unconventional
molecular-replacement procedure. Tens of putative models of
the C-terminal domain of the protein were constructed using a
multitude of homology-modelling algorithms, and these were
tested for the presence of signal in molecular-replacement
calculations. Of these, only the model calculated by the
SAM-T08 server gave a consistent and convincing solution,
but the resulting model was too inaccurate to allow phase
determination to proceed to completion. The application of
slow-cooling torsion-angle simulated annealing (started from
a very high temperature) drastically improved this initial
model to the point of allowing phasing through cycles of
model building and refinement to be initiated. The structure of
the protein is presented with emphasis on the presence of a
C(alpha)-modified proline at its active site, which was modelled as an
alpha-hydroxy-L-proline.
The structure of BC0361, a polysaccharide deacetylase from
Bacillus cereus, has been determined using an unconventional
molecular-replacement procedure. Tens of putative models of
the C-terminal domain of the protein were constructed using a
multitude of homology-modelling algorithms, and these were
tested for the presence of signal in molecular-replacement
calculations. Of these, only the model calculated by the
SAM-T08 server gave a consistent and convincing solution,
but the resulting model was too inaccurate to allow phase
determination to proceed to completion. The application of
slow-cooling torsion-angle simulated annealing (started from
a very high temperature) drastically improved this initial
model to the point of allowing phasing through cycles of
model building and refinement to be initiated. The structure of
the protein is presented with emphasis on the presence of a
C(alpha)-modified proline at its active site, which was modelled as an
alpha-hydroxy-L-proline.
33. Kokkinidis, M., Glykos, N.M. & Fadouloglou*, V.E. (2012), "Protein Flexibility and Enzymatic Catalysis", Adv. Protein Chem. Struct. Biol., 87, 181-218.
Electronic reprint (562 KBytes) : Local copy, or directly from Adv. Protein Chem. Struct. Biol., Copyright © Elsevier.

 The dynamic nature of protein structures has been recognized, established,
and accepted as an intrinsic fundamental property with major
consequences to their function. Nowadays, proteins are considered as
networks of continuous motions, which reflect local flexibility and a propensity
for global structural plasticity. Proteinâprotein and proteinâsmall
ligand interactions, signal transduction and assembly of macromolecular
machines, allosteric regulation and thermal enzymatic adaptation are
processes which require structural flexibility. In general, enzymes represent
an attractive class among proteins in the study of protein flexibility
and they can be used as model systems for understanding the implications
of protein fluctuations to biological function. Flexibility of the active site is
considered as a requirement for reduction of free energy barrier and
acceleration of the enzymatic reaction while there is growing evidence
which concerns the connection between flexibility and substrate turnover
rate. Moreover, the role of conformational flexibility has been well established
in connection with the accessibility of the active site, the binding of
substrates and ligands, and release of products, stabilization and trapping
of intermediates, orientation of the substrate into the binding cleft, adjustment
of the reaction environment, etc.
The dynamic nature of protein structures has been recognized, established,
and accepted as an intrinsic fundamental property with major
consequences to their function. Nowadays, proteins are considered as
networks of continuous motions, which reflect local flexibility and a propensity
for global structural plasticity. Proteinâprotein and proteinâsmall
ligand interactions, signal transduction and assembly of macromolecular
machines, allosteric regulation and thermal enzymatic adaptation are
processes which require structural flexibility. In general, enzymes represent
an attractive class among proteins in the study of protein flexibility
and they can be used as model systems for understanding the implications
of protein fluctuations to biological function. Flexibility of the active site is
considered as a requirement for reduction of free energy barrier and
acceleration of the enzymatic reaction while there is growing evidence
which concerns the connection between flexibility and substrate turnover
rate. Moreover, the role of conformational flexibility has been well established
in connection with the accessibility of the active site, the binding of
substrates and ligands, and release of products, stabilization and trapping
of intermediates, orientation of the substrate into the binding cleft, adjustment
of the reaction environment, etc.
32. Georgoulia, P.S. & Glykos*, N.M. (2011), "Using J-coupling constants for force field validation: Application to hepta-alanine", J. Phys. Chem. B, 115, 15221â15227.
Electronic reprint (3.6 MBytes) : Local copy, or directly from J. Phys. Chem. B, Copyright © American Chemical Society. See also the Supporting Information.
 A computational solution to the protein folding problem is the holy grail of
biomolecular simulation and of the corresponding force fields. The
complexity of the systems used for folding simulations precludes a direct
feedback between the simulations and the force fields, thus necessitating
the study of simpler systems with sufficient experimental data to allow
force field optimization and validation. Recent studies on short polyalanine
peptides of increasing length (up to penta-alanine) indicated the presence
of a systematic deviation between the experimental (NMR-derived) J-couplings
and the great majority of biomolecular force fields, with the Ï2 values for
even the best-performing force fields being in the 1.4â1.8 range. Here we
show that by increasing the number of residues to seven and by achieving
convergence through an increase of the simulation time to 2 ÎŒs, we can
identify one force field (the AMBER99SB force field, out of the three force
fields studied) which when compared with the experimental J-coupling data
(and for a specific set of Karplus-equation parameters and estimated
J-coupling errors previously used in the literature) gave a value of
Ï2=0.99, indicating that full statistical consistency between experiment and
simulation is feasible. However, and as a detailed analysis of the effects
of estimated errors shows, the Ï2 values may be unsuitable as indicators of
the goodness-of-fit of the various biomolecular force fields.
A computational solution to the protein folding problem is the holy grail of
biomolecular simulation and of the corresponding force fields. The
complexity of the systems used for folding simulations precludes a direct
feedback between the simulations and the force fields, thus necessitating
the study of simpler systems with sufficient experimental data to allow
force field optimization and validation. Recent studies on short polyalanine
peptides of increasing length (up to penta-alanine) indicated the presence
of a systematic deviation between the experimental (NMR-derived) J-couplings
and the great majority of biomolecular force fields, with the Ï2 values for
even the best-performing force fields being in the 1.4â1.8 range. Here we
show that by increasing the number of residues to seven and by achieving
convergence through an increase of the simulation time to 2 ÎŒs, we can
identify one force field (the AMBER99SB force field, out of the three force
fields studied) which when compared with the experimental J-coupling data
(and for a specific set of Karplus-equation parameters and estimated
J-coupling errors previously used in the literature) gave a value of
Ï2=0.99, indicating that full statistical consistency between experiment and
simulation is feasible. However, and as a detailed analysis of the effects
of estimated errors shows, the Ï2 values may be unsuitable as indicators of
the goodness-of-fit of the various biomolecular force fields.
31. Patapati, K.K. & Glykos*, N.M. (2011), "Three Force Fields' Views of the 310 Helix", Biophys. J., 101, 1766-1771.
Electronic reprint (805 KBytes) : Local copy, or directly from Biophys. J., 101, 1766-1771. Copyright © 2011 Biophysical Society.
 Slowly but steadily bibliographic evidence is accumulating that the apparent
convergence of the various biomolecular force fields as evidenced from
simulations of proteins in the folded state does not hold true for folding
simulations. Here we add one more example to the growing list of peptides
and proteins for which different force fields show irreconcilable
differences in their folding predictions, even at such a fundamental level
as that of a peptide's secondary structure. We show that for an undecamer
peptide that is known from two independent NMR structure determinations to
have a mainly 310-helical structure in solution, three mainstream
biomolecular force fields give completely disparate predictions: The CHARMM
force field (with the CMAP correction) predicts an outstandingly stable
α-helical structure, in disagreement not only with the experimental
structures, but also with experimental evidence obtained from circular
dichroism. OPLS-AA shows an almost totally disordered peptide with the most
frequently observed folded conformation corresponding to a β-hairpin-like
structure, again in disagreement with all available experimental evidence.
Only the AMBER99SB force field appears to qualitatively agree with not only
the general structural characteristics of the peptide (on the account of
both NMR- and CD-based experiments), but to also correctly predict some of
the experimentally observed interactions at the level of side chains.
Possible interpretations of these findings are discussed.
Slowly but steadily bibliographic evidence is accumulating that the apparent
convergence of the various biomolecular force fields as evidenced from
simulations of proteins in the folded state does not hold true for folding
simulations. Here we add one more example to the growing list of peptides
and proteins for which different force fields show irreconcilable
differences in their folding predictions, even at such a fundamental level
as that of a peptide's secondary structure. We show that for an undecamer
peptide that is known from two independent NMR structure determinations to
have a mainly 310-helical structure in solution, three mainstream
biomolecular force fields give completely disparate predictions: The CHARMM
force field (with the CMAP correction) predicts an outstandingly stable
α-helical structure, in disagreement not only with the experimental
structures, but also with experimental evidence obtained from circular
dichroism. OPLS-AA shows an almost totally disordered peptide with the most
frequently observed folded conformation corresponding to a β-hairpin-like
structure, again in disagreement with all available experimental evidence.
Only the AMBER99SB force field appears to qualitatively agree with not only
the general structural characteristics of the peptide (on the account of
both NMR- and CD-based experiments), but to also correctly predict some of
the experimentally observed interactions at the level of side chains.
Possible interpretations of these findings are discussed.
30. Glykos*, N.M. (2011), "On the application of structure-specific bulk-solvent models", Acta Crystallogr., D67, 739-741.
Electronic reprint (982 KBytes) : Local copy, or directly from Acta Cryst. D67, 739-741, Copyright © International Union of Crystallography.

 It is often discussed, mainly in connection with the rather high
macromolecular R factors, that the treatment of bulk solvent in
macromolecular refinement may lack the detail needed for modelling the
solvent environment of molecules as complex as proteins and nucleic acids.
This line of thought directly leads to the hypothesis that improvements in
the modelling of the bulk solvent may substantially improve the agreement
between the experimental data and the crystallographic models. Here, part of
this hypothesis is being tested through the construction, via
molecular-dynamics simulations, of a highly detailed, physics-based,
structure-specific and crystallographic data-agnostic model of the bulk
solvent of a known crystal structure. The water-distribution map obtained
from the simulation is converted (after imposing space-group symmetry) to a
constant (but scalable) partial structure factor which is then added in a
re-refinement of the crystal structure. Compared with the simple
Babinet-based correction, a reduction of the totally cross-validated free R
value by 0.3% is observed. The implications and possible interpretations of
these results are discussed.
It is often discussed, mainly in connection with the rather high
macromolecular R factors, that the treatment of bulk solvent in
macromolecular refinement may lack the detail needed for modelling the
solvent environment of molecules as complex as proteins and nucleic acids.
This line of thought directly leads to the hypothesis that improvements in
the modelling of the bulk solvent may substantially improve the agreement
between the experimental data and the crystallographic models. Here, part of
this hypothesis is being tested through the construction, via
molecular-dynamics simulations, of a highly detailed, physics-based,
structure-specific and crystallographic data-agnostic model of the bulk
solvent of a known crystal structure. The water-distribution map obtained
from the simulation is converted (after imposing space-group symmetry) to a
constant (but scalable) partial structure factor which is then added in a
re-refinement of the crystal structure. Compared with the simple
Babinet-based correction, a reduction of the totally cross-validated free R
value by 0.3% is observed. The implications and possible interpretations of
these results are discussed.
29. Glykos*, N.M. (2011), "The 11th Misconception?", A letter to the Editor, CBE Life Sci Educ, 10(1), 1-2.
Electronic reprint (123 KBytes) : Local copy, or directly from CBE Life Sci Educ, 10(1), 1-2, Copyright © Glykos under the Creative Commons Attribution License.
 Biological sequences are an abstraction of an abstraction: In the first
level, we substituted the complexity of a proper three-dimensional entity
(like an amino acid residue) with a two dimensional chemical formula
describing only composition and covalent bonding. In the second abstraction
layer, we substituted these chemical formulas with single alphabet letters.
And then we forgot about it, and started behaving as if sequences do exist,
as if this artificial one-dimensionality is real. Sequence usage became so
widespread, that not only we forgot that these one-dimensional strings of
letters do not (and never did) exist, but we have started using them for
dealing with problems (like protein folding) that by their nature defy this
whole 'sequence' abstraction. Maybe, just maybe, we have had more than
enough of 'sequences' ?
Biological sequences are an abstraction of an abstraction: In the first
level, we substituted the complexity of a proper three-dimensional entity
(like an amino acid residue) with a two dimensional chemical formula
describing only composition and covalent bonding. In the second abstraction
layer, we substituted these chemical formulas with single alphabet letters.
And then we forgot about it, and started behaving as if sequences do exist,
as if this artificial one-dimensionality is real. Sequence usage became so
widespread, that not only we forgot that these one-dimensional strings of
letters do not (and never did) exist, but we have started using them for
dealing with problems (like protein folding) that by their nature defy this
whole 'sequence' abstraction. Maybe, just maybe, we have had more than
enough of 'sequences' ?
28. Patapati, K.K. & Glykos*, N.M. (2010), "Order through Disorder: Hyper-Mobile C-Terminal Residues Stabilize the Folded State of a Helical Peptide. A Molecular Dynamics Study", PLoS ONE, 5, e15290.
Electronic reprint (607 KBytes) : Local copy, or directly from PLos ONE, 5, e15290, Copyright © Patapati & Glykos under the Creative Commons Attribution License, See also the Supporting Information.

 Conventional wisdom has it that the presence of disordered regions in the
three-dimensional structures of polypeptides not only does not contribute
significantly to the thermodynamic stability of their folded state, but, on
the contrary, that the presence of disorder leads to a decrease of the
corresponding proteins' stability. We have performed extensive 3.4 ÎŒs long
folding simulations (in explicit solvent and with full electrostatics) of an
undecamer peptide of experimentally known helical structure, both with and
without its disordered (four residue long) C-terminal tail. Our simulations
clearly indicate that the presence of the apparently disordered (in
structural terms) C-terminal tail, increases the thermodynamic stability of
the peptide's folded (helical) state. These results show that at least for
the case of relatively short peptides, the interplay between thermodynamic
stability and the apparent structural stability can be rather subtle, with
even disordered regions contributing significantly to the stability of the
folded state. Our results have clear implications for the understanding of
peptide energetics and the design of foldable peptides.
Conventional wisdom has it that the presence of disordered regions in the
three-dimensional structures of polypeptides not only does not contribute
significantly to the thermodynamic stability of their folded state, but, on
the contrary, that the presence of disorder leads to a decrease of the
corresponding proteins' stability. We have performed extensive 3.4 ÎŒs long
folding simulations (in explicit solvent and with full electrostatics) of an
undecamer peptide of experimentally known helical structure, both with and
without its disordered (four residue long) C-terminal tail. Our simulations
clearly indicate that the presence of the apparently disordered (in
structural terms) C-terminal tail, increases the thermodynamic stability of
the peptide's folded (helical) state. These results show that at least for
the case of relatively short peptides, the interplay between thermodynamic
stability and the apparent structural stability can be rather subtle, with
even disordered regions contributing significantly to the stability of the
folded state. Our results have clear implications for the understanding of
peptide energetics and the design of foldable peptides.
27. Fadouloglou, V.E., Stavrakoudis, S., Bouriotis, V., Kokkinidis, M., & Glykos*, N.M. (2009), "Molecular Dynamics Simulations of BcZBP, A Deacetylase from Bacillus cereus: Active Site Loops Determine Substrate Accessibility and Specificity", J. Chem. Theory Comput., 5, 3299-3311.
Electronic reprint (7.3 MBytes) : Local copy, or directly from J. Chem. Theory Comput., 5, 3299-3311, Copyright © American Chemical Society.
 BcZBP is an LmbE-like, homohexameric, zinc-dependent deacetylase from the
opportunistic pathogen Bacillus cereus with three, thus far uncharacterized,
homologues in B. anthracis. Although its specific substrate is still
unknown, the enzyme has been shown to preferentially deacetylate
N-acetylglucosamine and diacetylchitobiose via an active site based on a
zinc-binding motif of the type HXDDXnH. In the crystal structure, the active
site is located at a deep and partially blocked cleft formed at the
interface between monomers related by the molecular 3-fold axis, although
the major, in structural terms, building block of the enzyme is not the
trimer, but the intertwined dimer. Here, we report results from a 50 ns
molecular dynamics simulation of BcZBP in explicit solvent with full
electrostatics and show that (i) the view of the intertwined dimer as the
major structural and functional building block of this class of hexameric
enzymes is possibly an oversimplification of the rather complex dynamics
observed in the simulation, (ii) the most mobile (with respect to their
atomic fluctuations) parts of the structure coincide with three surface
loops surrounding the active site, and (iii) these mobile loops define the
active site's accessibility, and may be implicated in the determination of
the enzyme's specificity.
BcZBP is an LmbE-like, homohexameric, zinc-dependent deacetylase from the
opportunistic pathogen Bacillus cereus with three, thus far uncharacterized,
homologues in B. anthracis. Although its specific substrate is still
unknown, the enzyme has been shown to preferentially deacetylate
N-acetylglucosamine and diacetylchitobiose via an active site based on a
zinc-binding motif of the type HXDDXnH. In the crystal structure, the active
site is located at a deep and partially blocked cleft formed at the
interface between monomers related by the molecular 3-fold axis, although
the major, in structural terms, building block of the enzyme is not the
trimer, but the intertwined dimer. Here, we report results from a 50 ns
molecular dynamics simulation of BcZBP in explicit solvent with full
electrostatics and show that (i) the view of the intertwined dimer as the
major structural and functional building block of this class of hexameric
enzymes is possibly an oversimplification of the rather complex dynamics
observed in the simulation, (ii) the most mobile (with respect to their
atomic fluctuations) parts of the structure coincide with three surface
loops surrounding the active site, and (iii) these mobile loops define the
active site's accessibility, and may be implicated in the determination of
the enzyme's specificity.
26. Fadouloglou, V.E., Bastaki, M.N., Ashcroft, A.E., Phillips, S.E.V., Panopoulos, N.J., Glykos*, N.M., & Kokkinidis*, M. (2009), "On the quaternary association of the type III secretion system HrcQB-C protein: Experimental evidence differentiates among the various oligomerization models", J. Struct. Biol., 166, 214-225.
Electronic reprint (3.7 MBytes) : Local copy, or directly from J. Struct. Biol., 166, 214-225, Copyright © Elsevier B.V.
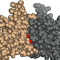
 The HrcQB protein from the plant pathogen Pseudomonas syringae is a core
component of the bacterial type III secretion apparatus. The core consists
of nine proteins widely conserved among animal and plant pathogens which
also share sequence and structural similarities with proteins from the
bacterial flagellum. Previous studies of the carboxy-terminal domain of
HrcQB (HrcQB-C) and its flagellar homologue, FliN-C, have revealed
extensive sequence and structural homologies, similar subcellular
localization, and participation in analogous protein-protein interaction
networks. It is not clear however whether the similarities between the two
proteins extend to the level of quaternary association which is essential
for the formation of higher-order structures within the TTSS. Even though
the crystal structure of the FliN is a dimer, more detailed studies support
a tetrameric donut-like association. However, both models, dimer and
donut-like tetramer, are quite different from the crystallographic elongated
dimer of dimers of the HrcQB-C. To resolve this discrepancy we performed a
multidisciplinary investigation of the quaternary association of the
HrcQB-C, including mass-spectrometry, electrophoresis in non-reductive
conditions, gel filtration, glutaraldehyde cross-linking and small angle
X-ray scattering. Our experiments indicate that stable tetramers of
elongated shape are assembled in solution, in agreement with the results of
crystallographic studies. Circular dichroism data are consistent with a
dimer-dimer interface analogous to the one established in the crystal
structure. Finally, molecular dynamics simulations reveal the relative
orientation of the dimers forming the tetramers and the possible differences
from that of the crystal structure.
The HrcQB protein from the plant pathogen Pseudomonas syringae is a core
component of the bacterial type III secretion apparatus. The core consists
of nine proteins widely conserved among animal and plant pathogens which
also share sequence and structural similarities with proteins from the
bacterial flagellum. Previous studies of the carboxy-terminal domain of
HrcQB (HrcQB-C) and its flagellar homologue, FliN-C, have revealed
extensive sequence and structural homologies, similar subcellular
localization, and participation in analogous protein-protein interaction
networks. It is not clear however whether the similarities between the two
proteins extend to the level of quaternary association which is essential
for the formation of higher-order structures within the TTSS. Even though
the crystal structure of the FliN is a dimer, more detailed studies support
a tetrameric donut-like association. However, both models, dimer and
donut-like tetramer, are quite different from the crystallographic elongated
dimer of dimers of the HrcQB-C. To resolve this discrepancy we performed a
multidisciplinary investigation of the quaternary association of the
HrcQB-C, including mass-spectrometry, electrophoresis in non-reductive
conditions, gel filtration, glutaraldehyde cross-linking and small angle
X-ray scattering. Our experiments indicate that stable tetramers of
elongated shape are assembled in solution, in agreement with the results of
crystallographic studies. Circular dichroism data are consistent with a
dimer-dimer interface analogous to the one established in the crystal
structure. Finally, molecular dynamics simulations reveal the relative
orientation of the dimers forming the tetramers and the possible differences
from that of the crystal structure.
25. Fadouloglou, V.E., Kokkinidis, M. & Glykos*, N.M. (2008), "Determination of protein oligomerization state: Two approaches based on glutaraldehyde crosslinking", Anal. Biochem., 373, 404-406.
Electronic reprint (605 KBytes) : Local copy, or directly from Anal. Biochem., 373, 404-406, Copyright © Elsevier B.V.

 Many biochemical and biophysical methods can be used to
characterize the oligomerization state of proteins. One of the most widely
used is glutaraldehyde crosslinking, mainly because of the minimum equipment
and reagents required. However, the crosslinking procedures currently in use
are impaired by the low specificity of the reagent, which can chemically
bond any two amino groups that are close in space. Thus, extensive and
time-consuming investigation of the reaction conditions is usually required.
Here we describe two approaches based on glutaraldehyde that readily give
reliable results.
Many biochemical and biophysical methods can be used to
characterize the oligomerization state of proteins. One of the most widely
used is glutaraldehyde crosslinking, mainly because of the minimum equipment
and reagents required. However, the crosslinking procedures currently in use
are impaired by the low specificity of the reagent, which can chemically
bond any two amino groups that are close in space. Thus, extensive and
time-consuming investigation of the reaction conditions is usually required.
Here we describe two approaches based on glutaraldehyde that readily give
reliable results.
24. Mizas, Ch., Sirakoulis, G.Ch., Mardiris, V., Karafyllidis, I., Glykos, N.M. & Sandaltzopoulos*, R. (2008), "Reconstruction of DNA sequences using genetic algorithms and cellular automata: Towards mutation prediction ?", BioSystems, 92, 61-68.
Electronic reprint (574 KBytes) : Local copy, or directly from BioSystems, 92, 61-68, Copyright © Elsevier B.V.

 Change of DNA sequence that fuels evolution is, to a certain
extent, a deterministic process because mutagenesis does not occur in an
absolutely random manner. So far, it has not been possible to decipher the
rules that govern DNA sequence evolution due to the extreme complexity of
the entire process. In our attempt to approach this issue we focus solely on
the mechanisms of mutagenesis and deliberately disregard the role of natural
selection. Hence, in this analysis, evolution refers to the accumulation of
genetic alterations that originate from mutations and are transmitted
through generations without being subjected to natural selection. We have
developed a software tool that allows modelling of a DNA sequence as a
one-dimensional cellular automaton (CA) with four states per cell which
correspond to the four DNA bases, i.e. A, C, T and G. The four states are
represented by numbers of the quaternary number system. Moreover, we have
developed genetic algorithms (GAs) in order to determine the rules of CA
evolution that simulate the DNA evolution process. Linear evolution rules
were considered and square matrices were used to represent them. If DNA
sequences of different evolution steps are available, our approach allows
the determination of the underlying evolution rule(s). Conversely, once the
evolution rules are deciphered, our tool may reconstruct the DNA sequence in
any previous evolution step for which the exact sequence information was
unknown. The developed tool may be used to test various parameters that
could influence evolution. We describe a paradigm relying on the assumption
that mutagenesis is governed by a near-neighbour-dependent mechanism. Based
on the satisfactory performance of our system in the deliberately
simplified example, we propose that our approach could offer a starting
point for future attempts to understand the mechanisms that govern
evolution. The developed software is open-source and has a user-friendly
graphical input interface.
Change of DNA sequence that fuels evolution is, to a certain
extent, a deterministic process because mutagenesis does not occur in an
absolutely random manner. So far, it has not been possible to decipher the
rules that govern DNA sequence evolution due to the extreme complexity of
the entire process. In our attempt to approach this issue we focus solely on
the mechanisms of mutagenesis and deliberately disregard the role of natural
selection. Hence, in this analysis, evolution refers to the accumulation of
genetic alterations that originate from mutations and are transmitted
through generations without being subjected to natural selection. We have
developed a software tool that allows modelling of a DNA sequence as a
one-dimensional cellular automaton (CA) with four states per cell which
correspond to the four DNA bases, i.e. A, C, T and G. The four states are
represented by numbers of the quaternary number system. Moreover, we have
developed genetic algorithms (GAs) in order to determine the rules of CA
evolution that simulate the DNA evolution process. Linear evolution rules
were considered and square matrices were used to represent them. If DNA
sequences of different evolution steps are available, our approach allows
the determination of the underlying evolution rule(s). Conversely, once the
evolution rules are deciphered, our tool may reconstruct the DNA sequence in
any previous evolution step for which the exact sequence information was
unknown. The developed tool may be used to test various parameters that
could influence evolution. We describe a paradigm relying on the assumption
that mutagenesis is governed by a near-neighbour-dependent mechanism. Based
on the satisfactory performance of our system in the deliberately
simplified example, we propose that our approach could offer a starting
point for future attempts to understand the mechanisms that govern
evolution. The developed software is open-source and has a user-friendly
graphical input interface.
23. Fadouloglou, V.E., Deli, A., Glykos, N.M., Psylinakis, E., Bouriotis, V. & Kokkinidis*, M. (2007), "Crystal structure of the BcZBP, a zinc-binding protein from Bacillus cereus. Functional insights from structural data", FEBS J., 274, 3044-3054.
Electronic reprint (1.2 MBytes) : Local copy, or directly from FEBS J., 274, 3044-3054, Copyright © Blackwell Publishing, Inc.

 Bacillus cereus is an opportunistic pathogenic bacterium closely related to
B. anthracis, the causative agent of anthrax in mammals. A
significant portion of the B. cereus chromosomal genes are common to
B. anthracis, including genes which in B. anthracis code for putative virulence
and surface proteins. B. cereus thus provides a convenient model organism
for studying proteins potentially associated with the pathogenicity of the
highly infectious B. anthracis. The zinc-binding protein of B. cereus,
BcZBP, is encoded from the bc1534 gene which has three homologues to
B. anthracis. The protein exhibits deacetylase activity with the N-acetyl
moiety of the N-acetylglucosamine and the diacetylchitobiose and
triacetylchitotriose. However, neither the specific substrate of the BcZBP
nor the biochemical pathway have been conclusively identified. Here, we
present the crystal structure of BcZBP at 1.8 A resolution. The
N-terminal part of the 234 amino acid protein adopts a Rossmann fold whereas
the C-terminal part consists of two beta strands and two alpha helices. In the
crystal, the protein forms a compact hexamer, in agreement with solution
data. A zinc binding site and a potential active site have been identified
in each monomer. These sites have extensive similarities to those found in
two known zinc-dependent hydrolases with deacetylase activity, MshB and
LpxC, despite a low degree of amino acid sequence identity. The functional
implications and a possible catalytic mechanism are discussed.
(2ixd.pdb).
Bacillus cereus is an opportunistic pathogenic bacterium closely related to
B. anthracis, the causative agent of anthrax in mammals. A
significant portion of the B. cereus chromosomal genes are common to
B. anthracis, including genes which in B. anthracis code for putative virulence
and surface proteins. B. cereus thus provides a convenient model organism
for studying proteins potentially associated with the pathogenicity of the
highly infectious B. anthracis. The zinc-binding protein of B. cereus,
BcZBP, is encoded from the bc1534 gene which has three homologues to
B. anthracis. The protein exhibits deacetylase activity with the N-acetyl
moiety of the N-acetylglucosamine and the diacetylchitobiose and
triacetylchitotriose. However, neither the specific substrate of the BcZBP
nor the biochemical pathway have been conclusively identified. Here, we
present the crystal structure of BcZBP at 1.8 A resolution. The
N-terminal part of the 234 amino acid protein adopts a Rossmann fold whereas
the C-terminal part consists of two beta strands and two alpha helices. In the
crystal, the protein forms a compact hexamer, in agreement with solution
data. A zinc binding site and a potential active site have been identified
in each monomer. These sites have extensive similarities to those found in
two known zinc-dependent hydrolases with deacetylase activity, MshB and
LpxC, despite a low degree of amino acid sequence identity. The functional
implications and a possible catalytic mechanism are discussed.
(2ixd.pdb).
22. Glykos*, N.M. (2007), "On the application of molecular-dynamics simulations to validate thermal parameters and to optimize TLS-group selection for macromolecular refinement", Acta Crystallogr., D63, 705-713.
Electronic reprint (1.4 MBytes) : Local copy, or directly from Acta Cryst. D63, 705-713, Copyright © International Union of Crystallography.
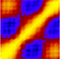
 Comparison of crystallographically determined and molecular dynamics
simulation-derived parameters for a small (26 kDa) homotetrameric
four-alpha-helical bundle protein revealed an unexpected pattern of similarities
and differences between experiment and simulation. On one hand, the protein
structure per se is exceptionally well preserved during the simulations,
with a root-mean-square deviation between the Ca atoms of the crystal
structure and the simulation-derived average structures of only 0.58 Angstrom,
which is not very different from the expected coordinate error of the
experimentally determined structure. On the other hand, comparison of the
temperature factors showed a large discrepancy, with the experimental B-factors
being approximately three times higher than the simulation-derived B-factors.
Closer examination of this discrepancy appears to validate the
molecular dynamics prediction and to implicate as its source static disorder
at the crystalline state, as indicated by the strong diffuse scattering and
pronounced anisotropy of the diffraction pattern of the protein crystals.
A posteriori re-refinement of the structure using a new TLS parameterization
scheme based on the results obtained from the simulations led to a further
reduction of the R factor and the free R value by 0.4% and 0.8%,
respectively, indicating that molecular-dynamics simulations have matured to
the point that they can be used to aid the selection of TLS groups for
macromolecular refinement.
Comparison of crystallographically determined and molecular dynamics
simulation-derived parameters for a small (26 kDa) homotetrameric
four-alpha-helical bundle protein revealed an unexpected pattern of similarities
and differences between experiment and simulation. On one hand, the protein
structure per se is exceptionally well preserved during the simulations,
with a root-mean-square deviation between the Ca atoms of the crystal
structure and the simulation-derived average structures of only 0.58 Angstrom,
which is not very different from the expected coordinate error of the
experimentally determined structure. On the other hand, comparison of the
temperature factors showed a large discrepancy, with the experimental B-factors
being approximately three times higher than the simulation-derived B-factors.
Closer examination of this discrepancy appears to validate the
molecular dynamics prediction and to implicate as its source static disorder
at the crystalline state, as indicated by the strong diffuse scattering and
pronounced anisotropy of the diffraction pattern of the protein crystals.
A posteriori re-refinement of the structure using a new TLS parameterization
scheme based on the results obtained from the simulations led to a further
reduction of the R factor and the free R value by 0.4% and 0.8%,
respectively, indicating that molecular-dynamics simulations have matured to
the point that they can be used to aid the selection of TLS groups for
macromolecular refinement.
21. Glykos*, N.M. (2006), "Carma: a molecular dynamics analysis program", J. Comput. Chem., 27, 1765-1768.
Electronic reprint (284 KBytes) : Local copy, or directly from Wiley InterScience, Copyright © Wiley InterScience.

 A computer program has been developed to aid the analysis of
molecular dynamics trajectories. The program is tuned for macromolecular
large-scale problems and supports features such as removal of global
translations-rotations of the solute, calculation of average distance maps
and their corresponding standard deviations, calculation of the
variance-covariance and cross-correlation matrices, and principal component
analysis of trajectories with the added ability to create artificial
trajectories based on selected eigenvectors. Limited graphics (trajectory
viewing) capabilities are also available.
A computer program has been developed to aid the analysis of
molecular dynamics trajectories. The program is tuned for macromolecular
large-scale problems and supports features such as removal of global
translations-rotations of the solute, calculation of average distance maps
and their corresponding standard deviations, calculation of the
variance-covariance and cross-correlation matrices, and principal component
analysis of trajectories with the added ability to create artificial
trajectories based on selected eigenvectors. Limited graphics (trajectory
viewing) capabilities are also available.
20. Glykos, N.M., Papanikolau, Y., Vlassi, M., Kotsifaki, D., Cesareni G. & Kokkinidis*, M. (2006), "Loopless Rop: Structure and Dynamics of an Engineered Homotetrameric Variant of the Repressor of Primer Protein", Biochemistry, 45, 10905-10919.
Electronic reprint (2.0 MBytes) : Local copy, or directly from the American Chemical Society, Copyright © American Chemical Society.

 The repressor of primer (Rop) protein has become a steady source
of surprises concerning the relationship between the sequences and the
structures of several of its mutants and variants. Here we add another piece
to the puzzle of Rop by showing that an engineered deletion mutant of the
protein (corresponding to a deletion of residues 30-34 of the wild-type
protein and designed to restore the heptad periodicity at the turn region)
results in a complete reorganization of the bundle which is converted from a
homodimer to a homotetramer. In contrast (and as previously shown), a
two-residue insertion, which also restores the heptad periodicity, is
essentially identical with wild-type Rop. The new deletion mutant structure
is a canonical, left-handed, all-antiparallel bundle with a completely
different hydrophobic core and distinct surface properties. The structure
agrees and qualitatively explains the results from functional,
thermodynamic, and kinetic studies which indicated that this deletion mutant
is a biologically inactive hyperstable homotetramer. Additional insight into
the stability and dynamics of the mutant structure has been obtained from
extensive molecular dynamics simulations in explicit water and with full
treatment of electrostatics.
(1qx8.pdb).
The repressor of primer (Rop) protein has become a steady source
of surprises concerning the relationship between the sequences and the
structures of several of its mutants and variants. Here we add another piece
to the puzzle of Rop by showing that an engineered deletion mutant of the
protein (corresponding to a deletion of residues 30-34 of the wild-type
protein and designed to restore the heptad periodicity at the turn region)
results in a complete reorganization of the bundle which is converted from a
homodimer to a homotetramer. In contrast (and as previously shown), a
two-residue insertion, which also restores the heptad periodicity, is
essentially identical with wild-type Rop. The new deletion mutant structure
is a canonical, left-handed, all-antiparallel bundle with a completely
different hydrophobic core and distinct surface properties. The structure
agrees and qualitatively explains the results from functional,
thermodynamic, and kinetic studies which indicated that this deletion mutant
is a biologically inactive hyperstable homotetramer. Additional insight into
the stability and dynamics of the mutant structure has been obtained from
extensive molecular dynamics simulations in explicit water and with full
treatment of electrostatics.
(1qx8.pdb).
19. Glykos*, N.M. (2005), "Qs v.1.3: a parallel version of Queen of Spades", J. Appl. Crystallogr., 38, 574-575.
Electronic reprint (65 KBytes) : Local copy, or directly from J. Appl. Crystallogr. 38, 574-575, Copyright © International Union of Crystallography.

 The program Queen of Spades encodes an algorithm for a stochastic multidimensional approach to
molecular replacement. The program has been shown to be capable of successfully
locating solutions even in cases as complex as a 23-dimensional, 4-body
problem. Recently, we extended our approach
to tackle the full molecular replacement problem by allowing the possibility
of using many different search models simultaneously, and showed that we
could successfully locate solutions in the case of a 17-dimensional problem
involving one DNA and two (different) protein search models.
This multimodel, multidimensional approach does have
its cost: with a few thousand unique reflections in a high symmetry space
group and with more than two search models, a typical Qs run would
take well over two to three weeks of CPU time on the fastest personal
workstations. The way forward for such computationally intensive
calculations is of course parallelisation. Here, I report the availability of
a parallel version of Qs which is based on the Message Passing
Interface (MPI) paradigm.
The program Queen of Spades encodes an algorithm for a stochastic multidimensional approach to
molecular replacement. The program has been shown to be capable of successfully
locating solutions even in cases as complex as a 23-dimensional, 4-body
problem. Recently, we extended our approach
to tackle the full molecular replacement problem by allowing the possibility
of using many different search models simultaneously, and showed that we
could successfully locate solutions in the case of a 17-dimensional problem
involving one DNA and two (different) protein search models.
This multimodel, multidimensional approach does have
its cost: with a few thousand unique reflections in a high symmetry space
group and with more than two search models, a typical Qs run would
take well over two to three weeks of CPU time on the fastest personal
workstations. The way forward for such computationally intensive
calculations is of course parallelisation. Here, I report the availability of
a parallel version of Qs which is based on the Message Passing
Interface (MPI) paradigm.
18. Glykos, N.M. & Kokkinidis*, M. (2004), "Structural Polymorphism of a Marginally Stable 4-alpha-Helical Bundle. Images of a Trapped Molten Globule ?", Proteins, 56, 420-425.
Electronic reprint (312 KBytes) : Local copy, or directly from Wiley InterScience, Copyright © Wiley InterScience.

 The Repressor of primer (Rop) protein is the paradigm of a canonical
(left-handed, all-anti-parallel) homodimeric 4-alpha-helical bundle. It
is known (through the analysis of an orthorhombic crystal form) that a
single alanine-to-proline amino-acid substitution at the turn region of Rop
(the A31P mutant) markedly changes the topology of the protein which is
converted to a right-handed, mixed parallel and antiparallel bundle. Here we
report the structure of this mutant in a second (monoclinic) crystal form
and show that although its topology remains unchanged, the differences
between the two A31P crystal structures are unexpectedly large, with a root
mean square deviation between equivalent Ca atoms of approximately
3A. Remarkably, a 3 ns molecular dynamics simulation of A31P which was
initiated from the orthorhombic form crystal structure, sampled an ensemble
of configurations rather similar to the structure seen in the monoclinic
form. This finding suggests that the observed crystal structures may
correspond to images of two conformers taken from a structurally
heterogeneous population of molecules at equilibrium. Comparison of the
A31P simulation with a 3 ns simulation of wild-type Rop indicated that the
mutant is an inherently highly flexible molecule, both with respect to the
relative placement of its helices and the malleability of its (loosely
packed) hydrophobic core. Based on these findings, we propose that the A31P
Rop mutant is an equilibrium molten globule and we attempt to interpret its
thermodynamic properties based on this assumption. Furthermore, we present
results from a 3 ns molecular dynamics simulation of a hypothetical
structure which was constructed by artificially mutating the alanine at
position 31 of the wild-type Rop structure to a proline. This hypothetical
A31P structure appears to be significantly more stable than the
experimentally determined one, leading us to propose that the observed A31P
structure may correspond to a kinetically trapped molten globule.
(1gmg.pdb).
The Repressor of primer (Rop) protein is the paradigm of a canonical
(left-handed, all-anti-parallel) homodimeric 4-alpha-helical bundle. It
is known (through the analysis of an orthorhombic crystal form) that a
single alanine-to-proline amino-acid substitution at the turn region of Rop
(the A31P mutant) markedly changes the topology of the protein which is
converted to a right-handed, mixed parallel and antiparallel bundle. Here we
report the structure of this mutant in a second (monoclinic) crystal form
and show that although its topology remains unchanged, the differences
between the two A31P crystal structures are unexpectedly large, with a root
mean square deviation between equivalent Ca atoms of approximately
3A. Remarkably, a 3 ns molecular dynamics simulation of A31P which was
initiated from the orthorhombic form crystal structure, sampled an ensemble
of configurations rather similar to the structure seen in the monoclinic
form. This finding suggests that the observed crystal structures may
correspond to images of two conformers taken from a structurally
heterogeneous population of molecules at equilibrium. Comparison of the
A31P simulation with a 3 ns simulation of wild-type Rop indicated that the
mutant is an inherently highly flexible molecule, both with respect to the
relative placement of its helices and the malleability of its (loosely
packed) hydrophobic core. Based on these findings, we propose that the A31P
Rop mutant is an equilibrium molten globule and we attempt to interpret its
thermodynamic properties based on this assumption. Furthermore, we present
results from a 3 ns molecular dynamics simulation of a hypothetical
structure which was constructed by artificially mutating the alanine at
position 31 of the wild-type Rop structure to a proline. This hypothetical
A31P structure appears to be significantly more stable than the
experimentally determined one, leading us to propose that the observed A31P
structure may correspond to a kinetically trapped molten globule.
(1gmg.pdb).
17. Papanikolau, Y., Kotsifaki, D., Fadouloglou, V.E., Gazi, A.D., Glykos, N.M., Cesareni G. & Kokkinidis*, M. (2004), "Ionic strength reducers: an efficient approach to protein purification and crystallization. Application to two Rop variants.", Acta Crystallogr., D60, 1334-1337.
Electronic reprint (539 KBytes) : Local copy, or directly from Acta Cryst. D60, 1334-1337, Copyright © International Union of Crystallography.
 Detailed knowledge of the influence of various parameters on
macromolecular solubility is essential for crystallization. The concept
of so-called ionic strength reducers provides insight into the changes
in solubility induced by organic solvents and hydrophilic polymers in
aqueous electrolytic solutions. A simple and efficient procedure is
presented which exploits the properties of ionic strength reducers in
the purification and crystallization of proteins. Using two designed
variants of the Rop protein as model systems, superior crystals have
been obtained compared with conventional techniques. This
procedure is particularly useful in cases where excessive nucleation
leads to the growth of a large number of tiny crystals that are useless
for crystallographic analysis.
Detailed knowledge of the influence of various parameters on
macromolecular solubility is essential for crystallization. The concept
of so-called ionic strength reducers provides insight into the changes
in solubility induced by organic solvents and hydrophilic polymers in
aqueous electrolytic solutions. A simple and efficient procedure is
presented which exploits the properties of ionic strength reducers in
the purification and crystallization of proteins. Using two designed
variants of the Rop protein as model systems, superior crystals have
been obtained compared with conventional techniques. This
procedure is particularly useful in cases where excessive nucleation
leads to the growth of a large number of tiny crystals that are useless
for crystallographic analysis.
16. Glykos*, N.M. & Kokkinidis, M. (2004), "Molecular Replacement with multiple different models", J. Appl. Crystallogr., 37, 159-161.
Electronic reprint (1.1 MBytes) : Local copy, or directly from J. Appl. Crystallogr. 37, 159-161, Copyright © International Union of Crystallography.
 Classical molecular replacement methods and the newer six-dimensional searches
treat molecular replacement as a succession of sub-problems of reduced dimensionality.
Due to their divide-and-conquer approach, these methods necessarily ignore (at
least during their early stages) the very knowledge that a target crystal structure may
comprise, for example, more than one copy of a search model, or, several models of
different types. We have previously described an algorithm for a stochastic multidimensional
molecular replacement search and showed that it can successfully locate
solutions even in cases as complex as a 23-dimensional, 4-body search. The original
description of the method only dealt with a special case of molecular replacement,
namely with the problem of placing n copies of only one search model in the
asymmetric unit of a target crystal structure. Here we present a natural generalisation of
this algorithm to deal with the full molecular replacement problem, that is, with the
problem of determining the orientations and positions of a total of n
copies of m different models which are assumed to be present in the asymmetric unit
of a target crystal structure. The generality of this approach is illustrated through its
successful application to a 17-dimensional, 3-model problem involving one DNA and
two protein molecules.
Classical molecular replacement methods and the newer six-dimensional searches
treat molecular replacement as a succession of sub-problems of reduced dimensionality.
Due to their divide-and-conquer approach, these methods necessarily ignore (at
least during their early stages) the very knowledge that a target crystal structure may
comprise, for example, more than one copy of a search model, or, several models of
different types. We have previously described an algorithm for a stochastic multidimensional
molecular replacement search and showed that it can successfully locate
solutions even in cases as complex as a 23-dimensional, 4-body search. The original
description of the method only dealt with a special case of molecular replacement,
namely with the problem of placing n copies of only one search model in the
asymmetric unit of a target crystal structure. Here we present a natural generalisation of
this algorithm to deal with the full molecular replacement problem, that is, with the
problem of determining the orientations and positions of a total of n
copies of m different models which are assumed to be present in the asymmetric unit
of a target crystal structure. The generality of this approach is illustrated through its
successful application to a 17-dimensional, 3-model problem involving one DNA and
two protein molecules.
15. Fadouloglou, V.E., Tampakaki, A.P., Glykos, N.M., Bastaki, N., Hadden, J.M., Phillips, S.E., Panopoulos, N.J. & Kokkinidis*, M. (2004), "Structure of HrcQb-C, a conserved component of the bacterial type III secretion systems", Proc. Natl. Acad. Sci. USA, 101, 70-75.
Electronic reprint (629 KBytes) : Local copy, or directly from Proc. Natl. Acad. Sci. USA, 101, 70-75, Copyright © National Academy of Sciences USA.

 Type III secretion systems enable plant and animal bacterial pathogens
to deliver virulence proteins into the cytosol of eukaryotic
host cells, causing a broad spectrum of diseases including
bacteremia, septicemia, typhoid fever, and bubonic plague in mammals,
and localized lesions, systemic wilting, and blights in plants. In
addition, type III secretion systems are also required for biogenesis
of the bacterial flagellum. The HrcQB protein, a component of the
secretion apparatus of Pseudomonas syringae with homologues in
all type III systems, has a variable N-terminal and a conserved
C-terminal domain (HrcQB-C). Here, we report the crystal structure
of HrcQB-C and show that this domain retains the ability of the
full-length protein to interact with other type III components. A 3D
analysis of sequence conservation patterns reveals two clusters of
residues potentially involved in proteinprotein interactions.
Based on the analogies between HrcQB and its flagellum homo-
logues, we propose that HrcQB-C participates in the formation of
a C-ring-like assembly.
(1o9y.pdb).
Type III secretion systems enable plant and animal bacterial pathogens
to deliver virulence proteins into the cytosol of eukaryotic
host cells, causing a broad spectrum of diseases including
bacteremia, septicemia, typhoid fever, and bubonic plague in mammals,
and localized lesions, systemic wilting, and blights in plants. In
addition, type III secretion systems are also required for biogenesis
of the bacterial flagellum. The HrcQB protein, a component of the
secretion apparatus of Pseudomonas syringae with homologues in
all type III systems, has a variable N-terminal and a conserved
C-terminal domain (HrcQB-C). Here, we report the crystal structure
of HrcQB-C and show that this domain retains the ability of the
full-length protein to interact with other type III components. A 3D
analysis of sequence conservation patterns reveals two clusters of
residues potentially involved in proteinprotein interactions.
Based on the analogies between HrcQB and its flagellum homo-
logues, we propose that HrcQB-C participates in the formation of
a C-ring-like assembly.
(1o9y.pdb).
14. Glykos*, N.M. & Kokkinidis, M. (2003), "Structure determination of a small protein through a 23-dimensional molecular replacement search", Acta Crystallogr., D59, 709-718.
Electronic reprint (3.8 MBytes) : Local copy, or directly from Acta Cryst. D59, 709-718, Copyright © International Union of Crystallography.

 The crystal structure of a 4-alpha-helical bundle protein has been
determined through the application of a 23-dimensional molecular replacement
search performed with a stochastic method. The search model for the
calculation was a 26 residue-long poly-alanine helix amounting to less than
13% of the total number of atoms in the asymmetric unit of the target
crystal structure. The crystal structure determination procedure is
presented in detail, with emphasis on the molecular replacement
calculations
(1gmg.pdb).
The crystal structure of a 4-alpha-helical bundle protein has been
determined through the application of a 23-dimensional molecular replacement
search performed with a stochastic method. The search model for the
calculation was a 26 residue-long poly-alanine helix amounting to less than
13% of the total number of atoms in the asymmetric unit of the target
crystal structure. The crystal structure determination procedure is
presented in detail, with emphasis on the molecular replacement
calculations
(1gmg.pdb).
13. Dennis, C.A., Glykos, N.M., Parsons, M.R. & Phillips*, S.E.V. (2002), "The structure of AhrC, the arginine repressor/activator protein from Bacillus subtilis", Acta Crystallogr., D58, 421-430.
Electronic reprint (1.6 MBytes) : Local copy, or directly from Acta Cryst. D58, 421-430, Copyright © International Union of Crystallography.

 In the gram positive bacterium Bacillus subtilis the concentration of the amino acid L-arginine is controlled
by the transcriptional regulator AhrC. The hexameric AhrC protein binds in an L-arginine-dependent manner to
pseudo-palindromic operators within the prometer regions of arginine biosynthesic and catabolic gene clusters.
AhrC binding results in the repression of transcription of biosynthetic genes and in the activation of transcription
of catabolic genes. We have determined the crystal structure of AhrC at 2.7A resulution. Each sununit of the protein
has two domains. The C-terminal domains are arranged with 32 point group symmetry and mediate the major inter-subunit
interactions. The N-terminal domains are located around this core, where they lie in weakly associated pairs but do not
obey strict symmetry. A structural comparison of AhrC with the arginine repressor from the thermophile
Bacillus stearothermophilus reveals close similarity in regions implicated in L-arginine binding and DNA recognition
but also some striking sequence differences, especially within the C-terminal oligomerisation domain, which may contribute
to the different thermostabilities of the proteins. Comparison of the crystal structure of AhrC with a 30A resolution
model obtained by combining X-ray structure factor amplitudes with phases derived from electron microscopic analyses of
AhrC crystals confirms the essential accuracy of the earlier model and suggests that such an approach may be more
widely useful for obtaining low resolution phase information
(1f9n.pdb).
In the gram positive bacterium Bacillus subtilis the concentration of the amino acid L-arginine is controlled
by the transcriptional regulator AhrC. The hexameric AhrC protein binds in an L-arginine-dependent manner to
pseudo-palindromic operators within the prometer regions of arginine biosynthesic and catabolic gene clusters.
AhrC binding results in the repression of transcription of biosynthetic genes and in the activation of transcription
of catabolic genes. We have determined the crystal structure of AhrC at 2.7A resulution. Each sununit of the protein
has two domains. The C-terminal domains are arranged with 32 point group symmetry and mediate the major inter-subunit
interactions. The N-terminal domains are located around this core, where they lie in weakly associated pairs but do not
obey strict symmetry. A structural comparison of AhrC with the arginine repressor from the thermophile
Bacillus stearothermophilus reveals close similarity in regions implicated in L-arginine binding and DNA recognition
but also some striking sequence differences, especially within the C-terminal oligomerisation domain, which may contribute
to the different thermostabilities of the proteins. Comparison of the crystal structure of AhrC with a 30A resolution
model obtained by combining X-ray structure factor amplitudes with phases derived from electron microscopic analyses of
AhrC crystals confirms the essential accuracy of the earlier model and suggests that such an approach may be more
widely useful for obtaining low resolution phase information
(1f9n.pdb).
12. Glykos*, N.M. & Kokkinidis, M. (2001), "Multidimensional Molecular Replacement", Acta Crystallogr., D57, 1462-1473.
Electronic reprint (1.9 MBytes) : Local copy, or directly from Acta Cryst. D57, 1462-1473, Copyright © International Union of Crystallography.
The ECM 20 (Krakow, 2001) presentation is also available for download (Powerpoint presentation, 1.6 MB).
 A method is described which attempts to simultaneously and independently
determine the positional and orientational parameters of all molecules
present in the asymmetric unit of a target crystal structure. This is
achieved through a reverse Monte Carlo optimisation of a suitable statistic
(like the R-factor or the linear correlation coefficient between the
observed and calculated amplitudes of the structure factors) in the
6n-dimensional space defined by the rotational and translational
parameters of the n search models. Results from the application of this
stochastic method ---obtained with a space group general computer program
which has been developed for this purpose--- indicate that with present-day
computing capabilities the method may successfully be applied to molecular
replacement problems for which the target crystal structure contains up to
three molecules per asymmetric unit. It is also shown that the method may be
useful in cases where the assumption of topological segregation of the self
and cross vectors in the Patterson function is violated (as may happen, for
example, in closely packed crystal structures).
A method is described which attempts to simultaneously and independently
determine the positional and orientational parameters of all molecules
present in the asymmetric unit of a target crystal structure. This is
achieved through a reverse Monte Carlo optimisation of a suitable statistic
(like the R-factor or the linear correlation coefficient between the
observed and calculated amplitudes of the structure factors) in the
6n-dimensional space defined by the rotational and translational
parameters of the n search models. Results from the application of this
stochastic method ---obtained with a space group general computer program
which has been developed for this purpose--- indicate that with present-day
computing capabilities the method may successfully be applied to molecular
replacement problems for which the target crystal structure contains up to
three molecules per asymmetric unit. It is also shown that the method may be
useful in cases where the assumption of topological segregation of the self
and cross vectors in the Patterson function is violated (as may happen, for
example, in closely packed crystal structures).
11. Fadouloglou, V.E., Glykos, N.M. & Kokkinidis*, M. (2001), "Side-chain conformations in 4-alpha-helical bundles", Protein Engng., 14, 321-328.
Electronic reprint (385 KBytes) : Local copy, or directly from Protein Engng. 14, 321-328, Copyright © Oxford University Press.
 The distribution of the chi1, chi2 dihedral angles in a data set consisting of
twelve unrelated 4-alpha-helical bundle proteins has been determined and
compared with that observed in globular proteins. The analysis suggests that
for this tertiary motif : (i) the side-chain conformations are limited to only
a subset of the conformations observed in globular proteins and are more
constrained that side chains in helical regions of globular proteins.
(ii) The side chains of Aspartic acid and Asparagine adopt occasionally a new,
topology-specific rotamer. (iii) The rotamer preferences of Tyrosine and
Isoleucine depend on whether they are located in the hydrophobic core of the
bundle or in a more exposed position. (iv) Naturally occuring mutations in
the hydrophobic core of 4-alpha-helical bundles follow a pattern that is
consistent with the notion that the mutated and wild-type residues have at
least one of their most highly populated rotamers in common. These findings
indicate a relationship between protein topology and preferred side-chains
conformations.
The distribution of the chi1, chi2 dihedral angles in a data set consisting of
twelve unrelated 4-alpha-helical bundle proteins has been determined and
compared with that observed in globular proteins. The analysis suggests that
for this tertiary motif : (i) the side-chain conformations are limited to only
a subset of the conformations observed in globular proteins and are more
constrained that side chains in helical regions of globular proteins.
(ii) The side chains of Aspartic acid and Asparagine adopt occasionally a new,
topology-specific rotamer. (iii) The rotamer preferences of Tyrosine and
Isoleucine depend on whether they are located in the hydrophobic core of the
bundle or in a more exposed position. (iv) Naturally occuring mutations in
the hydrophobic core of 4-alpha-helical bundles follow a pattern that is
consistent with the notion that the mutated and wild-type residues have at
least one of their most highly populated rotamers in common. These findings
indicate a relationship between protein topology and preferred side-chains
conformations.
10. Fadouloglou, V.E., Glykos*, N.M. & Kokkinidis, M. (2000), "A fast and inexpensive procedure for drying polyacrylamide gels.", Anal. Biochem., 287, 185-186.
Electronic reprint (69 KBytes) : Local copy, or directly from Anal. Biochem. 287, 185-186, Copyright © Academic Press.
 A simple procedure for drying polyacrylamide gels is described. This
only involves soaking the gel twice in low grade
ethanol : The gel is placed in a petri dish containing 5-10 gel volumes of low grade ethanol and stirred
for 10-15 minutes. After that time, the ethanol solution is refreshed and soaking continues for 5
minutes. During this second soak the gel becomes opaque, dehydrates and shrinks uniformly (without cracking)
by a factor of about 35-40%.
In the final step, the gel is removed from the ethanol solution,
placed on a hard (non-adhesive) surface, ethanol is allowed
to evaporate from its top surface, and then it is covered with a glass plate to avoid curling during
the final stages of ethanol evaporation. After few hours the gel is ready for storage.
A simple procedure for drying polyacrylamide gels is described. This
only involves soaking the gel twice in low grade
ethanol : The gel is placed in a petri dish containing 5-10 gel volumes of low grade ethanol and stirred
for 10-15 minutes. After that time, the ethanol solution is refreshed and soaking continues for 5
minutes. During this second soak the gel becomes opaque, dehydrates and shrinks uniformly (without cracking)
by a factor of about 35-40%.
In the final step, the gel is removed from the ethanol solution,
placed on a hard (non-adhesive) surface, ethanol is allowed
to evaporate from its top surface, and then it is covered with a glass plate to avoid curling during
the final stages of ethanol evaporation. After few hours the gel is ready for storage.
9. Spyridaki, A., Glykos, N.M., Kotsifaki, D., Fadouloglou, V. & Kokkinidis*, M. (2000), "Crystallization and diffraction to ultrahigh resolution (0.8A) of a designed variant of the Rop protein.", Acta Crystallogr., D56, 1015-1016.
Electronic reprint (237 KBytes) : Local copy, or directly from Acta Cryst. D56, 1015-1016, Copyright © International Union of Crystallography.

 The Rop protein is the paradigm of a highly regular 4-a-helix
bundle, and as such it has been subject to numerous structural
and mutagenesis studies. Crystals of a designed Rop variant which
establishes a continuous heptad pattern through the bend region
have been obtained by a combination of vapour diffusion and seeding
techniques. The crystals diffract to ultrahigh (0.8A)
resolution using synchrotron radiation and cryogenic conditions.
The Rop protein is the paradigm of a highly regular 4-a-helix
bundle, and as such it has been subject to numerous structural
and mutagenesis studies. Crystals of a designed Rop variant which
establishes a continuous heptad pattern through the bend region
have been obtained by a combination of vapour diffusion and seeding
techniques. The crystals diffract to ultrahigh (0.8A)
resolution using synchrotron radiation and cryogenic conditions.
8. Glykos, N.M. & Kokkinidis*, M. (2000), "On the distribution of the bulk solvent correction parameters", Acta Crystallogr., D56, 1070-1072.
Electronic reprint (384 KBytes) : Local copy, or directly from Acta Cryst. D56, 1070-1072, Copyright © International Union of Crystallography.

 The
distribution of the bulk solvent correction parameters (Bsol, ksol)
-as determined with an
exponential scaling algorithm based on Babinet's principle-
for 219 crystal
structures deposited with the Protein Data Bank is presented. The observed distribution
strongly suggests that the two parameters are not independent,
and a reasonable agreement
with the experimental data could be obtained through the application
of a simple exponential function.
Possible interpretations of this finding are discussed.
The
distribution of the bulk solvent correction parameters (Bsol, ksol)
-as determined with an
exponential scaling algorithm based on Babinet's principle-
for 219 crystal
structures deposited with the Protein Data Bank is presented. The observed distribution
strongly suggests that the two parameters are not independent,
and a reasonable agreement
with the experimental data could be obtained through the application
of a simple exponential function.
Possible interpretations of this finding are discussed.
7. Glykos*, N.M. & Kokkinidis, M. (2000), "GraphEnt : a maximum entropy program with graphics capabilities.", J. Appl. Crystallogr., 33, 982-985.
Electronic reprint (584 KBytes) : Local copy, or directly from J. Appl. Cryst. 33, 982-985, Copyright © International Union of Crystallography.

 A maximum entropy formalism aiming at the production of a
"maximally non-committal" map is a standard method in fields of
science like radioastronomy,
but a rare exception in both X-ray crystallography and electron microscopy
(or crystallography).
This is rather unfortunate, given the wealth of information that a MAXENT map can reveal,
especially when the
map itself
is the end product (for example, low resolution electron or potential density maps,
Patterson functions, deformation maps).
The program GraphEnt
attempts to automate the procedure of calculating
maximum entropy maps, with emphasis on the calculation of
difference Patterson functions
for macromolecular crystallographic problems, while
providing a useful graphical output of the current stage of the calculation.
A maximum entropy formalism aiming at the production of a
"maximally non-committal" map is a standard method in fields of
science like radioastronomy,
but a rare exception in both X-ray crystallography and electron microscopy
(or crystallography).
This is rather unfortunate, given the wealth of information that a MAXENT map can reveal,
especially when the
map itself
is the end product (for example, low resolution electron or potential density maps,
Patterson functions, deformation maps).
The program GraphEnt
attempts to automate the procedure of calculating
maximum entropy maps, with emphasis on the calculation of
difference Patterson functions
for macromolecular crystallographic problems, while
providing a useful graphical output of the current stage of the calculation.
6. Andreeva, A.E., Borissova, B.E., Mironova, R., Glykos, N.M., Kotsifaki, D., Ivanov, I., Krysteva, M. & Kokkinidis*, M. (2000), "Crystallization of type I chloramphenicol acetyltransferase : An approach based on the concept of ionic strength reducers", Acta Crystallogr., D56, 101-103.
Electronic reprint (319 KBytes) : Local copy, or directly from Acta Cryst. D56, 101-103, Copyright © International Union of Crystallography.

 Chloramphenicol acetyltransferase (CAT) is responsible for bacterial
resistance to chloramphenicol. It catalyzes inactivation of the
antibiotic by acetyl group transfer from acetyl CoA to one or
both hydroxyl groups of chloramphenicol. Type I CAT possesses
some unique properties which are not observed in other CAT variants.
Type I CAT overexpressed in E.coli was purified and crystals
with a resolution limit of 2.22 Å have been obtained
using a novel procedure which is based on the concept
of the "ionic strength reducers". The crystals have the symmetry
of spacegroup P1 and the unit cell dimensions are a=96.5, b=113.9,
c=114.2, alpha=119.9,
beta=94.1 and gamma=98.6 degrees. These
dimensions are consistent with four to six trimers per unit cell
corresponding to a solvent fraction ranging from 65 to 47%.
Chloramphenicol acetyltransferase (CAT) is responsible for bacterial
resistance to chloramphenicol. It catalyzes inactivation of the
antibiotic by acetyl group transfer from acetyl CoA to one or
both hydroxyl groups of chloramphenicol. Type I CAT possesses
some unique properties which are not observed in other CAT variants.
Type I CAT overexpressed in E.coli was purified and crystals
with a resolution limit of 2.22 Å have been obtained
using a novel procedure which is based on the concept
of the "ionic strength reducers". The crystals have the symmetry
of spacegroup P1 and the unit cell dimensions are a=96.5, b=113.9,
c=114.2, alpha=119.9,
beta=94.1 and gamma=98.6 degrees. These
dimensions are consistent with four to six trimers per unit cell
corresponding to a solvent fraction ranging from 65 to 47%.
5. Glykos*, N.M. & Kokkinidis, M. (2000), "A stochastic approach to Molecular Replacement", Acta Crystallogr., D56, 169-174.
Electronic reprint (415 KBytes) : Local copy, or directly from Acta Cryst. D56, 169-174, Copyright © International Union of Crystallography.
The CCP4 2001 Study Weekend presentation is also available for download (Powerpoint presentation, 1 MB).
The ECM 20 (Krakow, 2001) presentation is also available for download (Powerpoint presentation, 1.6 MB).

 The classical approach to the problem of placing n copies of a search
model in the asymmetric unit of a target crystal structure, is to divide
this 6n-dimensional optimisation problem into a succession of 3-dimensional
searches (rotation function followed by translation function searches for
each of the models). Here it is shown that a structure determination method
based on a reverse Monte Carlo minimisation of the conventional crystallographic
R-factor in the 6n-dimensional space defined by the rotational
and translational parameters of the n molecules, is both feasible
and practical, at least for small n. Because all parameters of all
molecules are determined simultaneously, this algorithm should improve
the signal-to-noise ratio in difficult cases involving high crystallographic/non-crystallographic
symmetry in tightly packed crystal forms. Preliminary results from the
application of this method -obtained with a space
group general computer program which has been developed for this purpose-
are presented.
The classical approach to the problem of placing n copies of a search
model in the asymmetric unit of a target crystal structure, is to divide
this 6n-dimensional optimisation problem into a succession of 3-dimensional
searches (rotation function followed by translation function searches for
each of the models). Here it is shown that a structure determination method
based on a reverse Monte Carlo minimisation of the conventional crystallographic
R-factor in the 6n-dimensional space defined by the rotational
and translational parameters of the n molecules, is both feasible
and practical, at least for small n. Because all parameters of all
molecules are determined simultaneously, this algorithm should improve
the signal-to-noise ratio in difficult cases involving high crystallographic/non-crystallographic
symmetry in tightly packed crystal forms. Preliminary results from the
application of this method -obtained with a space
group general computer program which has been developed for this purpose-
are presented.
4. Glykos, N.M. & Kokkinidis*, M. (1999), "Meaningful refinement of poly-alanine models using rigid-body simulated annealing : application to the structure determination of the A31P Rop mutant.", Acta Crystallogr., D55, 1301-1308.
Electronic reprint (1.7 MBytes) : Local copy, or directly from Acta Cryst. D55, 1301-1308, Copyright © International Union of Crystallography.

 Conventional least-squares or simulated annealing refinement of even correctly
positioned poly-alanine models of a target structure, results to a systematic
distortion of the molecular geometry and to a concomitant increase of the
mean phase difference from the correct phase set. Here it is shown that
iterative rigid-body simulated annealing refinement of poly-alanine models
employing successively fewer residues per rigid body (down to one alanine
residue per body) at a very high initial temperature (of the order of To=10000
K) and with the geometric energy terms switched on, not only preserves
the geometry of the model, but can also converge to an essentially correct
poly-alanine trace of the target structure even when the starting model
deviates systematically and significantly from the sought structure. As
an example of the application of the method we present details of the structure
determination of the alanine-31 to proline mutant of the Rop protein, where
an initial, roughly positioned poly-alanine model (giving an average phase
difference of 78.2 degrees from the final phase set) was successfully refined
against a 1.8A resolution native data set, leading to an essentially correct
model of the main chain with an average displacement of its atomic positions
from the final model of 0.275A. The phases calculated from this refined
poly-alanine model had an average difference of 43.8 degrees from the final
phase set (corresponding to a mean figure of merit of 0.72) and gave a
readily interpretable electron density map.
Conventional least-squares or simulated annealing refinement of even correctly
positioned poly-alanine models of a target structure, results to a systematic
distortion of the molecular geometry and to a concomitant increase of the
mean phase difference from the correct phase set. Here it is shown that
iterative rigid-body simulated annealing refinement of poly-alanine models
employing successively fewer residues per rigid body (down to one alanine
residue per body) at a very high initial temperature (of the order of To=10000
K) and with the geometric energy terms switched on, not only preserves
the geometry of the model, but can also converge to an essentially correct
poly-alanine trace of the target structure even when the starting model
deviates systematically and significantly from the sought structure. As
an example of the application of the method we present details of the structure
determination of the alanine-31 to proline mutant of the Rop protein, where
an initial, roughly positioned poly-alanine model (giving an average phase
difference of 78.2 degrees from the final phase set) was successfully refined
against a 1.8A resolution native data set, leading to an essentially correct
model of the main chain with an average displacement of its atomic positions
from the final model of 0.275A. The phases calculated from this refined
poly-alanine model had an average difference of 43.8 degrees from the final
phase set (corresponding to a mean figure of merit of 0.72) and gave a
readily interpretable electron density map.
3. Glykos*, N.M. (1999), "Pepinsky's Machine : an interactive, graphics-based Fourier synthesis program with applications in teaching and research.", J. Appl. Crystallogr., 32, 821-823.
Electronic reprint (660 KBytes) : Local copy, or directly from J. Appl. Cryst. 32, 821-823, FREE ITEM, Copyright © International Union of Crystallography.

 A computer program has been developed which
given a set of structure factor amplitudes for any centrosymmetric plane
group, displays the amplitude-weighted reciprocal lattice plane and allows
the user to interactively assign and modify the phases of the structure
factors, while observing the effect of these changes on the corresponding
electron density function. The program has the added feature of being able
to calculate and interactively display the electron density maps corresponding
to all phase combinations of a user-defined subset of structure factors.
The application of the program in both crystallographic teaching and research
are discussed.
A computer program has been developed which
given a set of structure factor amplitudes for any centrosymmetric plane
group, displays the amplitude-weighted reciprocal lattice plane and allows
the user to interactively assign and modify the phases of the structure
factors, while observing the effect of these changes on the corresponding
electron density function. The program has the added feature of being able
to calculate and interactively display the electron density maps corresponding
to all phase combinations of a user-defined subset of structure factors.
The application of the program in both crystallographic teaching and research
are discussed.
2. Glykos, N.M., Cesareni, G. & Kokkinidis*, M. (1999), "Protein plasticity to the extreme : Changing the topology of a 4-alpha-helical bundle with a single amino-acid substitution.", Structure 7, 597-603.
Electronic reprint (2.2 MBytes) : Local copy, or directly from ScienceDirect, Copyright © Current Biology Publications.
 The general experience from structural studies of single amino-acid substituted
mutant proteins, is that the effect of mutation is rather localised and
minor. Here we provide a counter-example to this statement by showing that
a single alanine to proline substitution in the turn region of a 4-alpha-helical
protein leads to a complete reorganisation of the whole molecule which
is converted from the canonical left-handed all-antiparallel form, to a
right-handed, mixed parallel and antiparallel bundle, which to the best
of our knowledge and belief represents a novel topological motif for this
class of proteins. Our results suggest a possible new mechanism for the
creation and evolution of protein folds, show the importance of the loop
regions in determining the allowable folding pathways and illustrate the
malleability of protein structure
(1b6q.pdb).
The general experience from structural studies of single amino-acid substituted
mutant proteins, is that the effect of mutation is rather localised and
minor. Here we provide a counter-example to this statement by showing that
a single alanine to proline substitution in the turn region of a 4-alpha-helical
protein leads to a complete reorganisation of the whole molecule which
is converted from the canonical left-handed all-antiparallel form, to a
right-handed, mixed parallel and antiparallel bundle, which to the best
of our knowledge and belief represents a novel topological motif for this
class of proteins. Our results suggest a possible new mechanism for the
creation and evolution of protein folds, show the importance of the loop
regions in determining the allowable folding pathways and illustrate the
malleability of protein structure
(1b6q.pdb).
1. Glykos, N.M., Holzenburg, A.K.H. & Phillips*, S.E.V. (1998), "Low resolution structural characterisation of the Arginine repressor/activator from Bacillus subtilis : A combined X-ray crystallographic and electron microscopical approach", Acta Crystallogr., D54, 215-225, and Acta Crystallogr., D54, 707-707.
Electronic reprint (2.6 MBytes) : Local copy, or directly from Acta Cryst. D54, 215-225, Copyright © International Union of Crystallography.

 Attempts to determine the X-ray crystal structure of the intact homohexameric arginine
repressor/activator from B. subtilis have so far been unsuccessful.
The major problem appears to be the lack of an isomorphous heavy atom derivative
with a manageable number of substitution sites. Here it is shown how electron
microscopy of thin three-dimensional crystals,
the same
as those used for the X-ray crystallographic
studies, made it possible (i) to obtain experimental support for some conclusions
drawn on the basis of X-ray data alone, (ii) to determine the low resolution
distribution of electron density in several different crystallographic
projections, and, (iii) to obtain a tentative low resolution model of the
whole hexamer.
Attempts to determine the X-ray crystal structure of the intact homohexameric arginine
repressor/activator from B. subtilis have so far been unsuccessful.
The major problem appears to be the lack of an isomorphous heavy atom derivative
with a manageable number of substitution sites. Here it is shown how electron
microscopy of thin three-dimensional crystals,
the same
as those used for the X-ray crystallographic
studies, made it possible (i) to obtain experimental support for some conclusions
drawn on the basis of X-ray data alone, (ii) to determine the low resolution
distribution of electron density in several different crystallographic
projections, and, (iii) to obtain a tentative low resolution model of the
whole hexamer.
0. Glykos, N.M. (1995), "Structural studies of the arginine repressor/activator from Bacillus subtilis", PhD thesis, Thesis advisor Prof Simon E.V. Phillips, Astbury Department of Biophysics, University of Leeds.
Electronic reprint (7.7 MBytes)

 In the presence of L-Arginine, AhrC --the Arginine-dependent
Repressor/Activator from Bacillus subtilis-- represses the
transcription of the genes encoding the anabolic and activates those
encoding the catabolic enzymes of arginine metabolism. AhrC is a homohexamer
of total molecular mass 105 kDa. It shows no homology to any of the
characterised DNA-binding motifs or DNA-binding proteins with the exception
of ArgR, the Arginine Repressor from Escherichia coli. ArgR does not
act as a transcription activator but it has been shown to be a necessary
accessory protein for the resolution --through site-specific
recombination-- of multimers of the ColE1 plasmid. Although the two
proteins share only 29% identity and are from such taxonomically distinct
prokaryotes, AhrC can complement E. coli ArgR- strains both in the
regulation of Arginine metabolism and the resolution of the ColE1 plasmid.
This thesis describes our attempts to determine the crystal structure of
AhrC. Three different crystal forms have been produced and characterised.
Useful derivatives have been prepared for two of these forms but the
determination of their heavy atom structures proved impossible. An attempt
to determine the low resolution structure of AhrC using Electron Microscopy
has been unsuccessful. Molecular Replacement using as a search model the
crystal structure of the hexameric core fragment of ArgR also failed to give
a convincing solution.
In the presence of L-Arginine, AhrC --the Arginine-dependent
Repressor/Activator from Bacillus subtilis-- represses the
transcription of the genes encoding the anabolic and activates those
encoding the catabolic enzymes of arginine metabolism. AhrC is a homohexamer
of total molecular mass 105 kDa. It shows no homology to any of the
characterised DNA-binding motifs or DNA-binding proteins with the exception
of ArgR, the Arginine Repressor from Escherichia coli. ArgR does not
act as a transcription activator but it has been shown to be a necessary
accessory protein for the resolution --through site-specific
recombination-- of multimers of the ColE1 plasmid. Although the two
proteins share only 29% identity and are from such taxonomically distinct
prokaryotes, AhrC can complement E. coli ArgR- strains both in the
regulation of Arginine metabolism and the resolution of the ColE1 plasmid.
This thesis describes our attempts to determine the crystal structure of
AhrC. Three different crystal forms have been produced and characterised.
Useful derivatives have been prepared for two of these forms but the
determination of their heavy atom structures proved impossible. An attempt
to determine the low resolution structure of AhrC using Electron Microscopy
has been unsuccessful. Molecular Replacement using as a search model the
crystal structure of the hexameric core fragment of ArgR also failed to give
a convincing solution.
Electronic reprint (1.6 MBytes) : Local copy, or directly from arXiv:2601.00618.

 We have previously shown that Good-Turing statistics can be applied to molecular
dynamics trajectories to estimate the probability of observing completely new
(thus far unobserved) biomolecular structures, and showed that the method is
stable, dependable and its predictions verifiable. The major problem with that
initial algorithm was the requirement for calculating and storing in memory the
two-dimensional RMSD matrix of the currently available trajectory. This
requirement precluded the application of the method to very long simulations.
Here we describe a new variant of the Good-Turing algorithm whose memory
requirements scale linearly with the number of structures in the trajectory,
making it suitable even for extremely long simulations. We show that the new
method gives essentially identical results with the older implementation, and
present results obtained from trajectories containing up to 22 million
structures. A computer program implementing the new algorithm is available from
standard repositories.
We have previously shown that Good-Turing statistics can be applied to molecular
dynamics trajectories to estimate the probability of observing completely new
(thus far unobserved) biomolecular structures, and showed that the method is
stable, dependable and its predictions verifiable. The major problem with that
initial algorithm was the requirement for calculating and storing in memory the
two-dimensional RMSD matrix of the currently available trajectory. This
requirement precluded the application of the method to very long simulations.
Here we describe a new variant of the Good-Turing algorithm whose memory
requirements scale linearly with the number of structures in the trajectory,
making it suitable even for extremely long simulations. We show that the new
method gives essentially identical results with the older implementation, and
present results obtained from trajectories containing up to 22 million
structures. A computer program implementing the new algorithm is available from
standard repositories.
55. Touliopoulos, S. & Glykos*, N.M. (2026), "Atomic density distributions in proteins: structural and functional implications", J. Mol. Graph. Model., 142, 109228.
Electronic reprint (9.0 MBytes) : Local copy, or directly from J. Mol. Graph. Model.. See also the Supporting information file.

 Atomic packing is an important metric for characterizing protein structures, as
it significantly influences various features including the stability, the rate
of evolution and the functional roles of proteins. Packing in protein structures
is a measure of the overall proximity between the proteins’ atoms and it can
vary notably among different structures. However, even single domain proteins do
not exhibit uniform packing throughout their structure. Protein cores in the
interior tend to be more tightly packed compared to the protein surface and the
presence of cavities and voids can disrupt that internal tight packing too.
Many different methods have been used to measure the quality of packing in
proteins, identify factors that influence it, and their possible implications.
In this work, we examine atomic density distributions derived from 21,255
non-redundant protein structures and show that statistically significant
differences between those distributions are present. The biomolecular assembly
unit was chosen as a representative for these structures. Addition of hydrogen
atoms and solvation was also performed to emulate a faithful representation of
the structures in vitro.
Several protein structures deviate significantly and systematically from the
average packing behavior. Hierarchical clustering indicated that there are
groups of structures with similar atomic density distributions. Search for
common features and patterns in these clusters showed that some of them include
proteins with characteristic structures such as coiled-coils and cytochromes.
Certain classification families such as hydrolases and transferases have also a
preference to appear more frequently in dense and loosely-packed clusters
respectively.
Regarding factors influencing packing, our results support knowledge that larger
structures have a smaller range in their density values, but tend to be more
loosely packed, compared to smaller proteins. We also used indicators, like
crystallographic water molecules abundance and B-factors as estimates of the
stability of the structures to reveal its relationship with packing.
Atomic packing is an important metric for characterizing protein structures, as
it significantly influences various features including the stability, the rate
of evolution and the functional roles of proteins. Packing in protein structures
is a measure of the overall proximity between the proteins’ atoms and it can
vary notably among different structures. However, even single domain proteins do
not exhibit uniform packing throughout their structure. Protein cores in the
interior tend to be more tightly packed compared to the protein surface and the
presence of cavities and voids can disrupt that internal tight packing too.
Many different methods have been used to measure the quality of packing in
proteins, identify factors that influence it, and their possible implications.
In this work, we examine atomic density distributions derived from 21,255
non-redundant protein structures and show that statistically significant
differences between those distributions are present. The biomolecular assembly
unit was chosen as a representative for these structures. Addition of hydrogen
atoms and solvation was also performed to emulate a faithful representation of
the structures in vitro.
Several protein structures deviate significantly and systematically from the
average packing behavior. Hierarchical clustering indicated that there are
groups of structures with similar atomic density distributions. Search for
common features and patterns in these clusters showed that some of them include
proteins with characteristic structures such as coiled-coils and cytochromes.
Certain classification families such as hydrolases and transferases have also a
preference to appear more frequently in dense and loosely-packed clusters
respectively.
Regarding factors influencing packing, our results support knowledge that larger
structures have a smaller range in their density values, but tend to be more
loosely packed, compared to smaller proteins. We also used indicators, like
crystallographic water molecules abundance and B-factors as estimates of the
stability of the structures to reveal its relationship with packing.
54. Vouzina, O.-D., Tafanidis, A. & Glykos*, N.M. (2024), "The curious case of A31P, a topology-switching mutant of the Repressor of Primer protein : A molecular dynamics study of its folding and misfolding", J. Chem. Inf. Model., 64, 6081-6091.
Electronic reprint (6.3 MBytes) : Local copy, or directly from J. Chem. Inf. Model.. See also the Supporting information file.

 The effect of mutations on protein structures is usually rather localized and
minor. Finding a mutation that can single-handedly change the fold and/or
topology of a protein structure is a rare exception. The A31P mutant of the
homodimeric Repressor of Primer (Rop) protein is one such exception: This single
mutation -and as demonstrated by two independent crystal structure
determinations- can convert the canonical (left-handed/all-antiparallel)
4-alpha-helical bundle of Rop, to a new form (right-handed/mixed parallel and
antiparallel bundle) displaying a previously unobserved 'bisecting U' topology.
The main problem with understanding the dramatic effect of this mutation on the
folding of Rop is to understand its very existence : Most computational methods
appear to agree that the mutation should have had no appreciable effect, with
the majority of energy minimization methods and protein structure prediction
protocols indicating that this mutation is fully consistent with the native Rop
structure, requiring only a local and minor change at the mutation site. Here we
use two long (10 us each) molecular dynamics simulations to compare the
stability and dynamics of the native Rop versus a hypothetical structure that is
identical with the native Rop but is carrying this single Alanine-31 to Proline
mutation. Comparative analysis of the two trajectories convincingly shows that
in contrast to the indications from energy minimization -but in agreement with
the experimental data-, this hypothetical native-like A31P structure is
unstable, with its turn regions almost completely unfolding, even under the
relatively mild 320K NpT simulations that we have used for this study. We
discuss the implication of these findings for the folding of the A31P mutant,
especially with respect to the proposed model of a double-funneled energy
landscape.
The effect of mutations on protein structures is usually rather localized and
minor. Finding a mutation that can single-handedly change the fold and/or
topology of a protein structure is a rare exception. The A31P mutant of the
homodimeric Repressor of Primer (Rop) protein is one such exception: This single
mutation -and as demonstrated by two independent crystal structure
determinations- can convert the canonical (left-handed/all-antiparallel)
4-alpha-helical bundle of Rop, to a new form (right-handed/mixed parallel and
antiparallel bundle) displaying a previously unobserved 'bisecting U' topology.
The main problem with understanding the dramatic effect of this mutation on the
folding of Rop is to understand its very existence : Most computational methods
appear to agree that the mutation should have had no appreciable effect, with
the majority of energy minimization methods and protein structure prediction
protocols indicating that this mutation is fully consistent with the native Rop
structure, requiring only a local and minor change at the mutation site. Here we
use two long (10 us each) molecular dynamics simulations to compare the
stability and dynamics of the native Rop versus a hypothetical structure that is
identical with the native Rop but is carrying this single Alanine-31 to Proline
mutation. Comparative analysis of the two trajectories convincingly shows that
in contrast to the indications from energy minimization -but in agreement with
the experimental data-, this hypothetical native-like A31P structure is
unstable, with its turn regions almost completely unfolding, even under the
relatively mild 320K NpT simulations that we have used for this study. We
discuss the implication of these findings for the folding of the A31P mutant,
especially with respect to the proposed model of a double-funneled energy
landscape.
53. Kolocouris*, A., Arkin, I. & Glykos*, N.M. (2022), "A proof-of-concept study of the secondary structure of influenza A, B M2 and MERS- and SARS-CoV E transmembrane peptides using folding molecular dynamics simulations in a membrane mimetic solvent", Phys. Chem. Chem. Phys., 24, 25391-25402.
Electronic reprint (1.8 MBytes) : Local copy, or directly from Phys. Chem. Chem. Phys., Copyright © Royal Society of Chemistry.

 We have carried out a proof-of-concept molecular dynamics (MD) simulation
with adaptive tempering in a membrane mimetic environment to study the folding
of single-pass membrane peptides. We tested the influenza A M2 viroporin,
influenza B M2 viroporin, and protein E from coronaviruses MERS-Cov-2 and
SARS-CoV-2 peptides with known experimental secondary structures in membrane
bilayers. The two influenza-derived peptides are significantly different in the
peptide sequence and secondary structure and more polar than the two
coronavirus-derived peptides. Through a total of more than 50 μs of simulation
time that could be accomplished in trifluoroethanol (TFE), as a membrane model,
we characterized comparatively the folding behavior, helical stability, and
helical propensity of these transmembrane peptides that match perfectly their
experimental secondary structures, and we identified common motifs that reflect
their quaternary organization and known (or not) biochemical function. We
showed that BM2 is organized into two structurally distinct parts: a
significantly more stable N-terminal half, and a fast-converting C-terminal
half that continuously folds and unfolds between α-helical structures and
non-canonical structures, which are mostly turns. In AM2, both the N-terminal
half and C-terminal half are very flexible. In contrast, the two
coronavirus-derived transmembrane peptides are much more stable and fast
helix-formers when compared with the influenza ones. In particular, the
SARS-derived peptide E appears to be the fastest and most stable helix-former
of all the four viral peptides studied, with a helical structure that persists
almost without disruption for the whole of its 10 μs simulation. By comparing
the results with experimental observations, we benchmarked TFE in studying the
conformation of membrane and hydrophobic peptides. This work provided accurate
results suggesting a methodology to run long MD simulations and predict
structural properties of biologically important membrane peptides.
We have carried out a proof-of-concept molecular dynamics (MD) simulation
with adaptive tempering in a membrane mimetic environment to study the folding
of single-pass membrane peptides. We tested the influenza A M2 viroporin,
influenza B M2 viroporin, and protein E from coronaviruses MERS-Cov-2 and
SARS-CoV-2 peptides with known experimental secondary structures in membrane
bilayers. The two influenza-derived peptides are significantly different in the
peptide sequence and secondary structure and more polar than the two
coronavirus-derived peptides. Through a total of more than 50 μs of simulation
time that could be accomplished in trifluoroethanol (TFE), as a membrane model,
we characterized comparatively the folding behavior, helical stability, and
helical propensity of these transmembrane peptides that match perfectly their
experimental secondary structures, and we identified common motifs that reflect
their quaternary organization and known (or not) biochemical function. We
showed that BM2 is organized into two structurally distinct parts: a
significantly more stable N-terminal half, and a fast-converting C-terminal
half that continuously folds and unfolds between α-helical structures and
non-canonical structures, which are mostly turns. In AM2, both the N-terminal
half and C-terminal half are very flexible. In contrast, the two
coronavirus-derived transmembrane peptides are much more stable and fast
helix-formers when compared with the influenza ones. In particular, the
SARS-derived peptide E appears to be the fastest and most stable helix-former
of all the four viral peptides studied, with a helical structure that persists
almost without disruption for the whole of its 10 μs simulation. By comparing
the results with experimental observations, we benchmarked TFE in studying the
conformation of membrane and hydrophobic peptides. This work provided accurate
results suggesting a methodology to run long MD simulations and predict
structural properties of biologically important membrane peptides.
52. Gkogka, I. & Glykos*, N.M. (2022), "Folding molecular dynamics simulation of T-peptide, a HIV viral entry inhibitor : Structure, dynamics, and comparison with the experimental data", J. Comput. Chem., 43, 942-952.
Electronic reprint (18 MBytes) : Local copy, or directly from J. Comput. Chem., Copyright © Wiley Periodicals, Inc.

 Peptide T is a synthetic octapeptide fragment, which corresponds to the region
185–192 of the gp120 HIV coat protein and functions as a viral entry inhibitor.
In this work, a folding molecular dynamics simulation of peptide T in a
membrane-mimicking (DMSO) solution was performed with the aim of characterizing
the peptide's structural and dynamical properties. We show that peptide T is
highly flexible and dynamic. The main structural characteristics observed were
rapidly interconverting short helical stretches and turns, with a notable
preference for the formation of β-turns. The simulation also indicated that the
C-terminal part appears to be more stable than the rest of the peptide, with the
most preferred conformation for residues 5–8 being a β-turn. In order to
validate the accuracy of the simulations, we compared our results with the
experimental NMR data obtained for the T-peptide in the same solvent. In
agreement with the simulation, the NMR data indicated the presence of a
preferred structure in solution that was consistent with a β-turn comprising the
four C-terminal residues. An additional comparison between the experimental and
simulation-derived chemical shifts also showed a reasonable agreement between
experiment and simulation, further validating the simulation-derived structural
characterization of the T-peptide. We conclude that peptide folding simulations
produce physically relevant results even when performed with organic solvents
that were not part of the force field parameterization procedure.
Peptide T is a synthetic octapeptide fragment, which corresponds to the region
185–192 of the gp120 HIV coat protein and functions as a viral entry inhibitor.
In this work, a folding molecular dynamics simulation of peptide T in a
membrane-mimicking (DMSO) solution was performed with the aim of characterizing
the peptide's structural and dynamical properties. We show that peptide T is
highly flexible and dynamic. The main structural characteristics observed were
rapidly interconverting short helical stretches and turns, with a notable
preference for the formation of β-turns. The simulation also indicated that the
C-terminal part appears to be more stable than the rest of the peptide, with the
most preferred conformation for residues 5–8 being a β-turn. In order to
validate the accuracy of the simulations, we compared our results with the
experimental NMR data obtained for the T-peptide in the same solvent. In
agreement with the simulation, the NMR data indicated the presence of a
preferred structure in solution that was consistent with a β-turn comprising the
four C-terminal residues. An additional comparison between the experimental and
simulation-derived chemical shifts also showed a reasonable agreement between
experiment and simulation, further validating the simulation-derived structural
characterization of the T-peptide. We conclude that peptide folding simulations
produce physically relevant results even when performed with organic solvents
that were not part of the force field parameterization procedure.
51. Mitsikas, D.A. & Glykos*, N.M. (2020), "A molecular dynamics simulation study on the propensity of Asn-Gly-containing heptapeptides towards β-turn structures: Comparison with ab initio quantum mechanical calculations", PLoS ONE, 15(12): e0243429.
Electronic reprint (2.8 MBytes) : Local copy, or directly from PLoS ONE, Copyright © Mitsikas & Glykos under the Creative Commons Attribution License.

 Both molecular mechanical and quantum mechanical calculations play an important
role in describing the behavior and structure of molecules. In this work, we
compare for the same peptide systems the results obtained from folding molecular
dynamics simulations with previously reported results from quantum mechanical
calculations. More specifically, three molecular dynamics simulations of 5 μs
each in explicit water solvent were carried out for three Asn-Gly-containing
heptapeptides, in order to study their folding and dynamics. Previous data,
based on quantum mechanical calculations within the DFT framework have shown
that these peptides adopt β-turn structures in aqueous solution, with type I’
β-turn being the most preferred motif. The results from our analyses indicate
that at least for the given systems, force field and simulation protocol, the
two methods diverge in their predictions. The possibility of a force
field-dependent deficiency is examined as a possible source of the observed
discrepancy.
Both molecular mechanical and quantum mechanical calculations play an important
role in describing the behavior and structure of molecules. In this work, we
compare for the same peptide systems the results obtained from folding molecular
dynamics simulations with previously reported results from quantum mechanical
calculations. More specifically, three molecular dynamics simulations of 5 μs
each in explicit water solvent were carried out for three Asn-Gly-containing
heptapeptides, in order to study their folding and dynamics. Previous data,
based on quantum mechanical calculations within the DFT framework have shown
that these peptides adopt β-turn structures in aqueous solution, with type I’
β-turn being the most preferred motif. The results from our analyses indicate
that at least for the given systems, force field and simulation protocol, the
two methods diverge in their predictions. The possibility of a force
field-dependent deficiency is examined as a possible source of the observed
discrepancy.
50. Kokkinidis, M., Glykos*, N.M. & Fadouloglou*, V.E. (2020), "Catalytic activity regulation through post-translational modification: the expanding universe of protein diversity", Adv. Protein Chem. Struct. Biol., 122, 97-125.
Electronic reprint (2.5 MBytes) : Local copy, or directly from Adv. Protein Chem. Struct. Biol., Copyright © Elsevier.

 Protein composition is restricted by the genetic code to a relatively small
number of natural amino acids. Similarly, the known three-dimensional structures
adopt a limited number of protein folds. However, proteins exert a large variety
of functions and show a remarkable ability for regulation and immediate response
to intracellular and extracellular stimuli. To some degree, the wide variability
of protein function can be attributed to the post-translational modifications.
Post-translational modifications have been observed in all kingdoms of life and
give to proteins a significant degree of chemical and consequently functional
and structural diversity. Their importance is partly reflected in the large
number of genes dedicated to their regulation. So far, hundreds of
post-translational modifications have been observed while it is believed that
many more are to be discovered along with the technological advances in
sequencing, proteomics, mass spectrometry and structural biology. Indeed, the
number of studies which report novel post translational modifications is getting
larger supporting the notion that their space is still largely unexplored. In
this review we explore the impact of post-translational modifications on protein
structure and function with emphasis on catalytic activity regulation. We
present examples of proteins and protein families whose catalytic activity is
substantially affected by the presence of post translational modifications and
we describe the molecular basis which underlies the regulation of the protein
function through these modifications. When available, we also summarize the
current state of knowledge on the mechanisms which introduce these modifications
to protein sites.
Protein composition is restricted by the genetic code to a relatively small
number of natural amino acids. Similarly, the known three-dimensional structures
adopt a limited number of protein folds. However, proteins exert a large variety
of functions and show a remarkable ability for regulation and immediate response
to intracellular and extracellular stimuli. To some degree, the wide variability
of protein function can be attributed to the post-translational modifications.
Post-translational modifications have been observed in all kingdoms of life and
give to proteins a significant degree of chemical and consequently functional
and structural diversity. Their importance is partly reflected in the large
number of genes dedicated to their regulation. So far, hundreds of
post-translational modifications have been observed while it is believed that
many more are to be discovered along with the technological advances in
sequencing, proteomics, mass spectrometry and structural biology. Indeed, the
number of studies which report novel post translational modifications is getting
larger supporting the notion that their space is still largely unexplored. In
this review we explore the impact of post-translational modifications on protein
structure and function with emphasis on catalytic activity regulation. We
present examples of proteins and protein families whose catalytic activity is
substantially affected by the presence of post translational modifications and
we describe the molecular basis which underlies the regulation of the protein
function through these modifications. When available, we also summarize the
current state of knowledge on the mechanisms which introduce these modifications
to protein sites.
49. Stylianakis, I., Shalev, A., Scheiner, S., Sigalas, M.P., Arkin, I.T., Glykos*, N.M. & Kolocouris*, A. (2020), "The balance between side‐chain and backbone‐driven association in folding of the α‐helical influenza A transmembrane peptide", J. Comput. Chem., 41, 2177-2188.
Electronic reprint (3.1 MBytes) : Local copy, or directly from J. Comput. Chem., Copyright © Wiley Periodicals, Inc.

 The correct balance between attractive, repulsive and peptide hydrogen bonding
interactions must be attained for proteins to fold correctly. To investigate
these important contributors, we sought a comparison of the folding between two
25‐residues peptides, the influenza A M2 protein transmembrane domain (M2TM) and
the 25‐Ala (Ala25). M2TM forms a stable α‐helix as is shown by circular
dichroism (CD) experiments. Molecular dynamics (MD) simulations with adaptive
tempering show that M2TM monomer is more dynamic in nature and quickly
interconverts between an ensemble of various α‐helical structures, and less
frequently turns and coils, compared to one α‐helix for Ala25. DFT calculations
suggest that folding from the extended structure to the α‐helical structure is
favored for M2TM compared with Ala25. This is due to CH⋯O attractive
interactions which favor folding to the M2TM α‐helix, and cannot be described
accurately with a force field. Using natural bond orbital (NBO) analysis and
quantum theory atoms in molecules (QTAIM) calculations, 26 CH⋯O interactions and
22 NH⋯O hydrogen bonds are calculated for M2TM. The calculations show that CH⋯O
hydrogen bonds, although individually weaker, have a cumulative effect that
cannot be ignored and may contribute as much as half of the total hydrogen
bonding energy, when compared to NH⋯O, to the stabilization of the α‐helix in
M2TM. Further, a strengthening of NH⋯O hydrogen bonding interactions is
calculated for M2TM compared to Ala25. Additionally, these weak CH⋯O
interactions can dissociate and associate easily leading to the ensemble of
folded structures for M2TM observed in folding MD simulations.
The correct balance between attractive, repulsive and peptide hydrogen bonding
interactions must be attained for proteins to fold correctly. To investigate
these important contributors, we sought a comparison of the folding between two
25‐residues peptides, the influenza A M2 protein transmembrane domain (M2TM) and
the 25‐Ala (Ala25). M2TM forms a stable α‐helix as is shown by circular
dichroism (CD) experiments. Molecular dynamics (MD) simulations with adaptive
tempering show that M2TM monomer is more dynamic in nature and quickly
interconverts between an ensemble of various α‐helical structures, and less
frequently turns and coils, compared to one α‐helix for Ala25. DFT calculations
suggest that folding from the extended structure to the α‐helical structure is
favored for M2TM compared with Ala25. This is due to CH⋯O attractive
interactions which favor folding to the M2TM α‐helix, and cannot be described
accurately with a force field. Using natural bond orbital (NBO) analysis and
quantum theory atoms in molecules (QTAIM) calculations, 26 CH⋯O interactions and
22 NH⋯O hydrogen bonds are calculated for M2TM. The calculations show that CH⋯O
hydrogen bonds, although individually weaker, have a cumulative effect that
cannot be ignored and may contribute as much as half of the total hydrogen
bonding energy, when compared to NH⋯O, to the stabilization of the α‐helix in
M2TM. Further, a strengthening of NH⋯O hydrogen bonding interactions is
calculated for M2TM compared to Ala25. Additionally, these weak CH⋯O
interactions can dissociate and associate easily leading to the ensemble of
folded structures for M2TM observed in folding MD simulations.
48. Riziotis, I.G. & Glykos*, N.M. (2019), "On the presence of short-range periodicities in protein structures that are not related to established secondary structure elements", Proteins, 87, 966-978.
Electronic reprint (5.7 MBytes) : Local copy, or directly from Proteins, Copyright © Wiley. See also the Supporting information file (2.3 Mbytes).

 Standard secondary structure elements such as α-helices or β-sheets, are characterized by repeating
backbone torsion angles (φ,ψ) at the single residue level. Two-residue motifs of the type (φ,ψ)₂ are
also observed in non-linear conformations, mainly turns. Taking these observations a step further, it
can be argued that there is no a priori reason why the presence of higher order periodicities can not
be envisioned in protein structures, such as, for example, periodic transitions between successive
residues of the type (...-α-β-α-β-α-...), or, (...-β-αL-β-αL-β-...), or, (...-α-β-αL-α-β-αL-...), etc., where
the symbols (α,β,αL) refer to the established Ramachandran-based residue conformations. From all
such possible higher order periodicities, here we examine the deposited (with the PDB) protein
structures for the presence of short-range periodical conformations comprising five consecutive
residues alternating between two (and only two) distinct Ramachandran regions, for example
conformations of the type (α-β-α-β-α) or (β-αL-β-αL-β) etc. Using a probabilistic approach we have
located several thousand of such peptapeptides, and these were clustered and analyzed in terms of
their structural characteristics, their sequences, and their putative functional correlations using a
gene ontology-based approach. We show that such non-standard short-range periodicities are
present in a large and functionally diverse sample of proteins, and can be grouped into two
structurally conserved major types. Examination of the structural context in which these
peptapeptides are observed gave no conclusive evidence for the presence of a persistent structural or
functional role of these higher order periodic conformations.
Standard secondary structure elements such as α-helices or β-sheets, are characterized by repeating
backbone torsion angles (φ,ψ) at the single residue level. Two-residue motifs of the type (φ,ψ)₂ are
also observed in non-linear conformations, mainly turns. Taking these observations a step further, it
can be argued that there is no a priori reason why the presence of higher order periodicities can not
be envisioned in protein structures, such as, for example, periodic transitions between successive
residues of the type (...-α-β-α-β-α-...), or, (...-β-αL-β-αL-β-...), or, (...-α-β-αL-α-β-αL-...), etc., where
the symbols (α,β,αL) refer to the established Ramachandran-based residue conformations. From all
such possible higher order periodicities, here we examine the deposited (with the PDB) protein
structures for the presence of short-range periodical conformations comprising five consecutive
residues alternating between two (and only two) distinct Ramachandran regions, for example
conformations of the type (α-β-α-β-α) or (β-αL-β-αL-β) etc. Using a probabilistic approach we have
located several thousand of such peptapeptides, and these were clustered and analyzed in terms of
their structural characteristics, their sequences, and their putative functional correlations using a
gene ontology-based approach. We show that such non-standard short-range periodicities are
present in a large and functionally diverse sample of proteins, and can be grouped into two
structurally conserved major types. Examination of the structural context in which these
peptapeptides are observed gave no conclusive evidence for the presence of a persistent structural or
functional role of these higher order periodic conformations.
47. Georgoulia, P.S. & Glykos*, N.M. (2019), "Molecular simulation of peptides coming of age: Accurate prediction of folding, dynamics and structures", Arch. Biochem. Biophys., 664, 76-88.
Electronic reprint (1.6 MBytes) : Local copy, or directly from Arch. Biochem. Biophys., Copyright © Elsevier Inc.

 The application of molecular dynamics simulations to study the folding and
dynamics of peptides has attracted a lot of interest in the last couple of
decades. Following the successful prediction of the folding of several proteins
using molecular simulation, foldable peptides emerged as a favourable system
mainly due to their application in improving protein structure prediction
methods and in drug design studies. However, their performance is inherently
linked to the accuracy of the empirical force fields used in the simulations,
whose optimisation and validation is of paramount importance. Here we review
the most important findings in the field of molecular peptide simulations and
highlight the significant advancements made over the last twenty years. Special
reference is made on the simulation of disordered peptides and the remaining
challenge to find a force field able to describe accurately their
conformational landscape.
The application of molecular dynamics simulations to study the folding and
dynamics of peptides has attracted a lot of interest in the last couple of
decades. Following the successful prediction of the folding of several proteins
using molecular simulation, foldable peptides emerged as a favourable system
mainly due to their application in improving protein structure prediction
methods and in drug design studies. However, their performance is inherently
linked to the accuracy of the empirical force fields used in the simulations,
whose optimisation and validation is of paramount importance. Here we review
the most important findings in the field of molecular peptide simulations and
highlight the significant advancements made over the last twenty years. Special
reference is made on the simulation of disordered peptides and the remaining
challenge to find a force field able to describe accurately their
conformational landscape.
46. Georgoulia*, P.S. & Glykos, N.M. (2018), "Folding Molecular Dynamics Simulation of a gp41-Derived Peptide Reconcile Divergent Structure Determinations", ACS Omega, 3, 14746-14754.
Electronic reprint (1.7 MBytes) : Local copy, or directly from ACS Omega, Copyright © American Chemical Society. See also the Supporting information file (11 Mbytes).

 T-20 peptide is the first FDA-approved fusion inhibitor against AIDS/HIV-1 gp41
protein. Part of it, the gp41[659-671] peptide, that contains the complete
epitope for the neutralizing 2F5 monoclonal antibody, has been found
experimentally in a number of divergent structures. Herein, we attempt to
reconcile them by using unbiased large-scale all-atom molecular dynamics
folding simulations. We show that our approach can successfully capture the
peptide's heterogeneity and reach each and every experimentally determined
conformation in sub-angstrom accuracy, whilst preserving the peptide's
disordered nature. Our analysis also unveils that the minor refinements within
the AMBER99SB family of force fields can lead to appreciable differences in the
predicted conformational stability arising from subtle differences in the
helical- and β-region of the Ramachandran plot. Our work underlines the
contribution of molecular dynamics simulation in structurally characterizing
pharmacologically important peptides of ambiguous structure.
T-20 peptide is the first FDA-approved fusion inhibitor against AIDS/HIV-1 gp41
protein. Part of it, the gp41[659-671] peptide, that contains the complete
epitope for the neutralizing 2F5 monoclonal antibody, has been found
experimentally in a number of divergent structures. Herein, we attempt to
reconcile them by using unbiased large-scale all-atom molecular dynamics
folding simulations. We show that our approach can successfully capture the
peptide's heterogeneity and reach each and every experimentally determined
conformation in sub-angstrom accuracy, whilst preserving the peptide's
disordered nature. Our analysis also unveils that the minor refinements within
the AMBER99SB family of force fields can lead to appreciable differences in the
predicted conformational stability arising from subtle differences in the
helical- and β-region of the Ramachandran plot. Our work underlines the
contribution of molecular dynamics simulation in structurally characterizing
pharmacologically important peptides of ambiguous structure.
45. Adamidou, T., Arvaniti, K.-O. & Glykos*, N.M. (2018), "Folding Simulations of a Nuclear Receptor Box-Containing Peptide Demonstrate the Structural Persistence of the LxxLL Motif Even in the Absence of Its Cognate Receptor", J. Phys. Chem. B, 122, 106â116.
Electronic reprint (2.5 MBytes) : Local copy, or directly from J. Phys. Chem. B, Copyright © American Chemical Society. See also the Supporting information file (3.3 Mbytes).

 Regulation of nuclear receptors by their coactivators involves the
recognition and binding of a specific sequence motif contained in the
coactivator sequence. This motif is known as the nuclear receptor (NR) box
and contains a conserved LxxLL subsequence, where L is leucine and x is any
amino acid residue. Crystallographic studies have shown that the LxxLL
motifs adopt an α-helical conformation when bound to their cognate nuclear
receptors. Here we use an extensive set of folding molecular dynamics
simulations to examine whether the α-helical conformation demonstrated by
the LxxLL motifs in the bound state may represent a persistent structural
preference of these peptides even in the absence of their cognate receptors.
To this end, we have performed a grand total of 35 ÎŒs of adaptive tempering
folding simulations of an NR-box-containing peptide derived from
Drosophila's fushi tarazu segmentation gene product. Our
simulationsâperformed using full electrostatics and an explicit
representation of two different solvents (water and a TFE/water
mixture)âclearly indicate the presence of a persistent helical preference of
the LxxLL motif with a concomitant native-like structure and contacts
between the motif's leucine residues. To lend further support to our
findings, we compare the simulation-derived peptide dynamics with
experimental NMR-derived nuclear Overhauser effect (NOE) measurements that
had been previously obtained for the same peptide in the same two solvents.
The comparison demonstrates a quantitative agreement between simulation and
experiment with average upper bound NOE violations of less than 0.084 Ã
,
thus independently validating our main conclusion concerning the intrinsic
preference of NR-box motifs to form helical structures even in the absence
of their cognate receptors.
Regulation of nuclear receptors by their coactivators involves the
recognition and binding of a specific sequence motif contained in the
coactivator sequence. This motif is known as the nuclear receptor (NR) box
and contains a conserved LxxLL subsequence, where L is leucine and x is any
amino acid residue. Crystallographic studies have shown that the LxxLL
motifs adopt an α-helical conformation when bound to their cognate nuclear
receptors. Here we use an extensive set of folding molecular dynamics
simulations to examine whether the α-helical conformation demonstrated by
the LxxLL motifs in the bound state may represent a persistent structural
preference of these peptides even in the absence of their cognate receptors.
To this end, we have performed a grand total of 35 ÎŒs of adaptive tempering
folding simulations of an NR-box-containing peptide derived from
Drosophila's fushi tarazu segmentation gene product. Our
simulationsâperformed using full electrostatics and an explicit
representation of two different solvents (water and a TFE/water
mixture)âclearly indicate the presence of a persistent helical preference of
the LxxLL motif with a concomitant native-like structure and contacts
between the motif's leucine residues. To lend further support to our
findings, we compare the simulation-derived peptide dynamics with
experimental NMR-derived nuclear Overhauser effect (NOE) measurements that
had been previously obtained for the same peptide in the same two solvents.
The comparison demonstrates a quantitative agreement between simulation and
experiment with average upper bound NOE violations of less than 0.084 Ã
,
thus independently validating our main conclusion concerning the intrinsic
preference of NR-box motifs to form helical structures even in the absence
of their cognate receptors.
44. Fadouloglou, V.E., Balomenou, S., Aivaliotis, M., Kotsifaki, D., Arnaouteli, S., Tomatsidou, A., Efstathiou, G., Kountourakis, N., Miliara, S., Griniezaki, M., Tsalafouta, A., Pergantis, S.A., Boneca, I.G., Glykos, N.M., Bouriotis, V. & Kokkinidis*, M. (2017), "An unusual α-carbon hydroxylation of proline promotes active-site maturation", J. Am. Chem. Soc., 139, 5330â5337.
Electronic reprint (2.5 MBytes) : Local copy, or directly from J. Am. Chem. Soc., Copyright © American Chemical Society.

 The full extent of proline (Pro) hydroxylation has yet to be established, as it is largely
unexplored in bacteria. We describe here a so far unknown Pro hydroxylation activity which occurs in
active sites of polysaccharide deacetylases (PDAs) from bacterial pathogens, modifying the protein
backbone at the Cα atom of a Pro residue to produce 2-hydroxyproline (2-Hyp). This process modifies with
high specificity a conserved Pro, shares with the deacetylation reaction the same active site and one catalytic
residue and utilizes molecular oxygen as source for the hydroxyl group oxygen of 2-Hyp. By
providing additional hydrogen bonding capacity, the Pro â 2-Hyp conversion alters the active site and
enhances significantly deacetylase activity, probably by creating a more favorable environment for
transition state stabilization. Our results classify this process as an active site "maturation", which is
highly atypical by being a protein backbone modifying activity, rather than a side-chain modifying one.
The full extent of proline (Pro) hydroxylation has yet to be established, as it is largely
unexplored in bacteria. We describe here a so far unknown Pro hydroxylation activity which occurs in
active sites of polysaccharide deacetylases (PDAs) from bacterial pathogens, modifying the protein
backbone at the Cα atom of a Pro residue to produce 2-hydroxyproline (2-Hyp). This process modifies with
high specificity a conserved Pro, shares with the deacetylation reaction the same active site and one catalytic
residue and utilizes molecular oxygen as source for the hydroxyl group oxygen of 2-Hyp. By
providing additional hydrogen bonding capacity, the Pro â 2-Hyp conversion alters the active site and
enhances significantly deacetylase activity, probably by creating a more favorable environment for
transition state stabilization. Our results classify this process as an active site "maturation", which is
highly atypical by being a protein backbone modifying activity, rather than a side-chain modifying one.
43. Serafeim A.-P., Salamanos, G., Patapati, K.K. & Glykos*, N.M. (2016), "Sensitivity of Folding Molecular Dynamics Simulations to Even Minor Force Field Changes", J. Chem. Inf. Model., 56, 2035-2041.
Electronic reprint (2.6 MBytes) : Local copy, or directly from J. Chem. Inf. Model., Copyright © American Chemical Society. See also the Supporting information file.

 We examine the sensitivity of folding molecular dynamics simulations on the
choice between three variants of the same force field (the AMBER99SB force
field and its ILDN, NMR-ILDN and STAR-ILDN variants). Using two different
peptide systems (a marginally stable helical peptide and a β-hairpin) and a
grand total of more than 20 ÎŒs of simulation time we show that even
relatively minor force field changes can lead to appreciable differences in
the peptide folding behavior.
We examine the sensitivity of folding molecular dynamics simulations on the
choice between three variants of the same force field (the AMBER99SB force
field and its ILDN, NMR-ILDN and STAR-ILDN variants). Using two different
peptide systems (a marginally stable helical peptide and a β-hairpin) and a
grand total of more than 20 ÎŒs of simulation time we show that even
relatively minor force field changes can lead to appreciable differences in
the peptide folding behavior.
42. Baltzis, A.S. & Glykos*, N.M. (2016), "Characterizing a partially ordered miniprotein through folding molecular dynamics simulations: Comparison with the experimental data", Prot. Sci., 25, 587â596.
Electronic reprint (3.3 MBytes) : Local copy, or directly from Protein Science, Copyright © Wiley Periodicals, Inc.

 The villin headpiece helical subdomain (HP36) is one of the best known model
systems for computational studies of fast-folding all-α miniproteins. HP21
is a peptide fragment âderived from HP36â comprising only the first and
second helices of the full domain. Experimental studies showed that although
HP21 is mostly unfolded in solution, it does maintain some persistent
native-like structure as indicated by the analysis of NMR-derived chemical
shifts. Here we compare the experimental data for HP21 with the results
obtained from a 15 ÎŒs long folding molecular dynamics simulation performed
in explicit water and with full electrostatics. We find that the simulation
is in good agreement with the experiment and faithfully reproduces the major
experimental findings, namely that (a) HP21 is disordered in solution with
less that 10% of the trajectory corresponding to transiently stable
structures, (b) the most highly populated conformer is a native-like
structure with an RMSD from the corresponding portion of the HP36 crystal
structure of less than 1Ã
, (c) the simulation-derived chemical shifts âover
the whole length of the trajectoryâ are in reasonable agreement with the
experiment giving reduced Ï2 values of 1.6, 1.4 and 0.8 for the ÎÎŽ13Cα,
ÎÎŽ13CO and ÎÎŽ13Cβ secondary shifts respectively (becoming 0.8, 0.7, and 0.3
when only the major peptide conformer is considered), and finally, (d) the
secondary structure propensity scores are in very good agreement with the
experiment and clearly indicate the higher stability of the first helix. We
conclude that folding molecular dynamics simulations can be a useful tool
for the structural characterization of even marginally stable peptides.
The villin headpiece helical subdomain (HP36) is one of the best known model
systems for computational studies of fast-folding all-α miniproteins. HP21
is a peptide fragment âderived from HP36â comprising only the first and
second helices of the full domain. Experimental studies showed that although
HP21 is mostly unfolded in solution, it does maintain some persistent
native-like structure as indicated by the analysis of NMR-derived chemical
shifts. Here we compare the experimental data for HP21 with the results
obtained from a 15 ÎŒs long folding molecular dynamics simulation performed
in explicit water and with full electrostatics. We find that the simulation
is in good agreement with the experiment and faithfully reproduces the major
experimental findings, namely that (a) HP21 is disordered in solution with
less that 10% of the trajectory corresponding to transiently stable
structures, (b) the most highly populated conformer is a native-like
structure with an RMSD from the corresponding portion of the HP36 crystal
structure of less than 1Ã
, (c) the simulation-derived chemical shifts âover
the whole length of the trajectoryâ are in reasonable agreement with the
experiment giving reduced Ï2 values of 1.6, 1.4 and 0.8 for the ÎÎŽ13Cα,
ÎÎŽ13CO and ÎÎŽ13Cβ secondary shifts respectively (becoming 0.8, 0.7, and 0.3
when only the major peptide conformer is considered), and finally, (d) the
secondary structure propensity scores are in very good agreement with the
experiment and clearly indicate the higher stability of the first helix. We
conclude that folding molecular dynamics simulations can be a useful tool
for the structural characterization of even marginally stable peptides.
41. Baltzis, A.S., Koukos, P.I. & Glykos*, N.M. (2015), "Clustering of molecular dynamics trajectories via peak-picking in multidimensional PCA-derived distributions", arXiv:1512.04024 [q-bio.BM].
Electronic reprint (826 KBytes) : Local copy, or directly from arXiv:1512.04024.

 We describe a robust, fast, and memory-efficient procedure that can cluster
millions of structures derived from molecular dynamics simulations. The
essence of the method is based on a peak-picking algorithm applied to three-
and five-dimensional distributions of the principal components derived from
the trajectory and automatically supports both Cartesian and dihedral
PCA-based clustering. The density threshold required for identifying
isolated peaks (which correspond to discrete clusters) is determined through
the application of a variance-explained criterion which allows for a
completely automated clustering procedure with no user intervention. In this
communication we describe the algorithm and present some of the results
obtained from the application of the method as implemented in the molecular
dynamics analysis programs carma, grcarma. and cluster5D. We conclude with a
discussion of the limitations and possible pitfalls of this method.
We describe a robust, fast, and memory-efficient procedure that can cluster
millions of structures derived from molecular dynamics simulations. The
essence of the method is based on a peak-picking algorithm applied to three-
and five-dimensional distributions of the principal components derived from
the trajectory and automatically supports both Cartesian and dihedral
PCA-based clustering. The density threshold required for identifying
isolated peaks (which correspond to discrete clusters) is determined through
the application of a variance-explained criterion which allows for a
completely automated clustering procedure with no user intervention. In this
communication we describe the algorithm and present some of the results
obtained from the application of the method as implemented in the molecular
dynamics analysis programs carma, grcarma. and cluster5D. We conclude with a
discussion of the limitations and possible pitfalls of this method.
40. Koukos, P.I. & Glykos*, N.M. (2014), "Folding Molecular Dynamics Simulations Accurately Predict the Effect of Mutations on the Stability and Structure of a Vammin-Derived Peptide", J. Phys. Chem. B, 118, 10076â10084.
Electronic reprint (3.7 MBytes) : Local copy, or directly from J. Phys. Chem. B, Copyright © American Chemical Society. See also the Supporting Information.

 Folding molecular dynamics simulations amounting to a grand total of 4 ÎŒs of
simulation time were performed on two peptides (with native and mutated
sequences) derived from loop 3 of the vammin protein and the results
compared with the experimentally known peptide stabilities and structures.
The simulations faithfully and accurately reproduce the major experimental
findings and show that (a) the native peptide is mostly disordered in
solution, (b) the mutant peptide has a well-defined and stable structure,
and (c) the structure of the mutant is an irregular β-hairpin with a
non-glycine β-bulge, in excellent agreement with the peptideâs known NMR
structure. Additionally, the simulations also predict the presence of a very
small β-hairpin-like population for the native peptide but surprisingly
indicate that this population is structurally more similar to the structure
of the native peptide as observed in the vammin protein than to the NMR
structure of the isolated mutant peptide. We conclude that, at least for the
given system, force field, and simulation protocol, folding molecular
dynamics simulations appear to be successful in reproducing the
experimentally accessible physical reality to a satisfactory level of detail
and accuracy.
Folding molecular dynamics simulations amounting to a grand total of 4 ÎŒs of
simulation time were performed on two peptides (with native and mutated
sequences) derived from loop 3 of the vammin protein and the results
compared with the experimentally known peptide stabilities and structures.
The simulations faithfully and accurately reproduce the major experimental
findings and show that (a) the native peptide is mostly disordered in
solution, (b) the mutant peptide has a well-defined and stable structure,
and (c) the structure of the mutant is an irregular β-hairpin with a
non-glycine β-bulge, in excellent agreement with the peptideâs known NMR
structure. Additionally, the simulations also predict the presence of a very
small β-hairpin-like population for the native peptide but surprisingly
indicate that this population is structurally more similar to the structure
of the native peptide as observed in the vammin protein than to the NMR
structure of the isolated mutant peptide. We conclude that, at least for the
given system, force field, and simulation protocol, folding molecular
dynamics simulations appear to be successful in reproducing the
experimentally accessible physical reality to a satisfactory level of detail
and accuracy.
39. Koukos, P.I. & Glykos*, N.M. (2014), "On the application of Good-Turing statistics to quantify convergence of biomolecular simulations", J. Chem. Inf. Model., 54, 209-217.
Electronic reprint (1.1 MBytes) : Local copy, or directly from J. Chem. Inf. Model., Copyright © American Chemical Society.

 Quantifying convergence and sufficient sampling of macromolecular molecular
dynamics simulations is more often than not a source of controversy (and of
various ad hoc solutions) in the field. Clearly, the only reasonable,
consistent and satisfying way to infer convergence (or otherwise) of a
molecular dynamics trajectory must be based on probability theory. Ideally,
the question we would wish to answer is the following : "What is the
probability that a molecular configuration important for the analysis in
hand has not yet been observed ?". Here we propose a method for answering a
variant of this question by using the Good-Turing formalism for frequency
estimation of unobserved species in a sample. Although several approaches
may be followed in order to deal with the problem of discretizing the
configurational space, for this work we use the classical RMSD matrix as a
means to answering the following question : "What is the probability that a
molecular configuration with an RMSD (from all other already observed
configurations) higher than a given threshold has not actually been observed
?". We apply the proposed method to several different trajectories and show
that the procedure appears to be both computationally stable and internally
consistent. A free, open-source program implementing these ideas is
immediately available for download via public repositories.
Quantifying convergence and sufficient sampling of macromolecular molecular
dynamics simulations is more often than not a source of controversy (and of
various ad hoc solutions) in the field. Clearly, the only reasonable,
consistent and satisfying way to infer convergence (or otherwise) of a
molecular dynamics trajectory must be based on probability theory. Ideally,
the question we would wish to answer is the following : "What is the
probability that a molecular configuration important for the analysis in
hand has not yet been observed ?". Here we propose a method for answering a
variant of this question by using the Good-Turing formalism for frequency
estimation of unobserved species in a sample. Although several approaches
may be followed in order to deal with the problem of discretizing the
configurational space, for this work we use the classical RMSD matrix as a
means to answering the following question : "What is the probability that a
molecular configuration with an RMSD (from all other already observed
configurations) higher than a given threshold has not actually been observed
?". We apply the proposed method to several different trajectories and show
that the procedure appears to be both computationally stable and internally
consistent. A free, open-source program implementing these ideas is
immediately available for download via public repositories.
38. Koukos, P.I. & Glykos*, N.M. (2013), "grcarma: A Fully Automated Task-Oriented Interface for the Analysis of Molecular Dynamics Trajectories", J. Comput. Chem., 34, 2310-2312, Cover story for Vol.34, Issue 26.
Electronic reprint (929 KBytes) : Local copy, or directly from J. Comput. Chem., Copyright © Wiley Periodicals, Inc.

 We report the availability of grcarma, a program encoding for
a fully automated set of tasks aiming to simplify the analysis
of molecular dynamics trajectories of biological macromolecules.
It is a cross-platform, Perl/Tk-based front-end to the program
carma and is designed to facilitate the needs of the
novice as well as those of the expert user, while at the same
time maintaining a user-friendly and intuitive design. Particular
emphasis was given to the automation of several tedious
tasks, such as extraction of clusters of structures based on
dihedral and Cartesian principal component analysis, secondary
structure analysis, calculation and display of root-meansquare
deviation (RMSD) matrices, calculation of entropy, calculation
and analysis of varianceâcovariance matrices, calculation
of the fraction of native contacts, etc. The program is
free-open source software available immediately for download.
We report the availability of grcarma, a program encoding for
a fully automated set of tasks aiming to simplify the analysis
of molecular dynamics trajectories of biological macromolecules.
It is a cross-platform, Perl/Tk-based front-end to the program
carma and is designed to facilitate the needs of the
novice as well as those of the expert user, while at the same
time maintaining a user-friendly and intuitive design. Particular
emphasis was given to the automation of several tedious
tasks, such as extraction of clusters of structures based on
dihedral and Cartesian principal component analysis, secondary
structure analysis, calculation and display of root-meansquare
deviation (RMSD) matrices, calculation of entropy, calculation
and analysis of varianceâcovariance matrices, calculation
of the fraction of native contacts, etc. The program is
free-open source software available immediately for download.
37. Kontopoulos, D.-G. & Glykos*, N.M. (2013), "Pinda: A Web service for detection and analysis of intraspecies gene duplication events", Comput. Methods Programs Biomed., 111, 711-714.
Electronic reprint (729 KBytes) : Local copy, or directly from Comput. Methods Programs Biomed., Copyright © Elsevier Ltd.

 We present Pinda, a Web service for the detection and analysis of possible
duplications of a given protein or DNA sequence within a source species.
Pinda fully automates the whole gene duplication detection procedure, from
performing the initial similarity searches, to generating the multiple
sequence alignments and the corresponding phylogenetic trees,
to bootstrapping the trees and producing a Z-score-based list of duplication
candidates for the input sequence. Pinda has been cross-validated using an
extensive set of known and bibliographically characterized duplication
events. The service facilitates the automatic and dependable identification
of gene duplication events, using some of the most successful bioinformatics
software to perform an extensive analysis protocol. Pinda will prove of
use for the analysis of newly discovered genes and proteins, thus also
assisting the study of recently sequenced genomes. The service's location is
http://orion.mbg.duth.gr/Pinda. The source code is freely available via
https://github.com/dgkontopoulos/Pinda/.
We present Pinda, a Web service for the detection and analysis of possible
duplications of a given protein or DNA sequence within a source species.
Pinda fully automates the whole gene duplication detection procedure, from
performing the initial similarity searches, to generating the multiple
sequence alignments and the corresponding phylogenetic trees,
to bootstrapping the trees and producing a Z-score-based list of duplication
candidates for the input sequence. Pinda has been cross-validated using an
extensive set of known and bibliographically characterized duplication
events. The service facilitates the automatic and dependable identification
of gene duplication events, using some of the most successful bioinformatics
software to perform an extensive analysis protocol. Pinda will prove of
use for the analysis of newly discovered genes and proteins, thus also
assisting the study of recently sequenced genomes. The service's location is
http://orion.mbg.duth.gr/Pinda. The source code is freely available via
https://github.com/dgkontopoulos/Pinda/.
36. Georgoulia, P.S. & Glykos*, N.M. (2013), "On the Foldability of Tryptophan-Containing Tetra- and Pentapeptides: An Exhaustive Molecular Dynamics Study", J. Phys. Chem. B, 117, 5522â5532.
Electronic reprint (3.1 MBytes) : Local copy, or directly from J. Phys. Chem. B, Copyright © American Chemical Society. See also the Supporting Information.

 Short peptides serve as minimal model systems
to decipher the determinants of foldability due to their
simplicity arising from their smaller size, their ability to echo
protein-like structural characteristics, and their direct implication
in force field validation. Here, we describe an effort to
identify small peptides that can still form stable structures in
aqueous solutions. We followed the in silico folding of a
selected set of 8640 tryptophan-containing tetra- and
pentapeptides through 15 210 molecular dynamics simulations
amounting to a total of 272.46 ÎŒs using explicit representation
of the solute and full treatment of the electrostatics. The
evaluation and sorting of peptides is achieved through scoring
functions, which include terms based on interatomic vector distances, atomic
fluctuations, and rmsd matrices between successive
frames of a trajectory. Highly scored peptides are studied further via
successive simulation rounds of increasing simulation length
and using different empirical force fields. Our method suggested only a
handful of peptides with strong foldability prognosis. The
discrepancies between the predictions of the various force fields for such
short sequences are also extensively discussed. We
conclude that the vast majority of such short peptides do not adopt
significantly stable structures in water solutions, at least based
on our computational predictions. The present work can be utilized in the
rational design and engineering of bioactive peptides
with desired molecular properties.
Short peptides serve as minimal model systems
to decipher the determinants of foldability due to their
simplicity arising from their smaller size, their ability to echo
protein-like structural characteristics, and their direct implication
in force field validation. Here, we describe an effort to
identify small peptides that can still form stable structures in
aqueous solutions. We followed the in silico folding of a
selected set of 8640 tryptophan-containing tetra- and
pentapeptides through 15 210 molecular dynamics simulations
amounting to a total of 272.46 ÎŒs using explicit representation
of the solute and full treatment of the electrostatics. The
evaluation and sorting of peptides is achieved through scoring
functions, which include terms based on interatomic vector distances, atomic
fluctuations, and rmsd matrices between successive
frames of a trajectory. Highly scored peptides are studied further via
successive simulation rounds of increasing simulation length
and using different empirical force fields. Our method suggested only a
handful of peptides with strong foldability prognosis. The
discrepancies between the predictions of the various force fields for such
short sequences are also extensively discussed. We
conclude that the vast majority of such short peptides do not adopt
significantly stable structures in water solutions, at least based
on our computational predictions. The present work can be utilized in the
rational design and engineering of bioactive peptides
with desired molecular properties.
35. Patmanidis, I. & Glykos*, N.M. (2013), "As good as it gets? Folding molecular dynamics simulations of the LytA choline-binding peptide result to an exceptionally accurate model of the peptide structure", J. Mol. Graph. Model., 41, 68-71.
Electronic reprint (686 KBytes) : Local copy, or directly from J. Mol. Graph. Model., 41, 68-71, Copyright © Elsevier Inc.

 Folding simulations of a choline-binding peptide derived from the Streptococcus pneumoniae LytA protein
converged to a model of the peptide's folded state structure which is in outstanding agreement with the
experimentally-determined structures, reaching values for the root mean squared deviation as low as
0.24 Ã
for the peptide's backbone atoms and 0.65 Ã
for all non-hydrogen atoms.
Folding simulations of a choline-binding peptide derived from the Streptococcus pneumoniae LytA protein
converged to a model of the peptide's folded state structure which is in outstanding agreement with the
experimentally-determined structures, reaching values for the root mean squared deviation as low as
0.24 Ã
for the peptide's backbone atoms and 0.65 Ã
for all non-hydrogen atoms.
34. Fadouloglou, V.E., Kapanidou, M., Agiomirgianaki, A., Arnaouteli, S., Bouriotis, V., Glykos*, N.M. & Kokkinidis*, M. (2013), "Structure determination through homology modelling and torsion-angle simulated annealing: application to a polysaccharide deacetylase from Bacillus cereus", Acta Crystallogr., D69, 276-283.
Electronic reprint (2.4 MBytes) : Local copy, or directly from Acta Crystallogr., D69, 276-283, Copyright © International Union of Crystallography.

 The structure of BC0361, a polysaccharide deacetylase from
Bacillus cereus, has been determined using an unconventional
molecular-replacement procedure. Tens of putative models of
the C-terminal domain of the protein were constructed using a
multitude of homology-modelling algorithms, and these were
tested for the presence of signal in molecular-replacement
calculations. Of these, only the model calculated by the
SAM-T08 server gave a consistent and convincing solution,
but the resulting model was too inaccurate to allow phase
determination to proceed to completion. The application of
slow-cooling torsion-angle simulated annealing (started from
a very high temperature) drastically improved this initial
model to the point of allowing phasing through cycles of
model building and refinement to be initiated. The structure of
the protein is presented with emphasis on the presence of a
C(alpha)-modified proline at its active site, which was modelled as an
alpha-hydroxy-L-proline.
The structure of BC0361, a polysaccharide deacetylase from
Bacillus cereus, has been determined using an unconventional
molecular-replacement procedure. Tens of putative models of
the C-terminal domain of the protein were constructed using a
multitude of homology-modelling algorithms, and these were
tested for the presence of signal in molecular-replacement
calculations. Of these, only the model calculated by the
SAM-T08 server gave a consistent and convincing solution,
but the resulting model was too inaccurate to allow phase
determination to proceed to completion. The application of
slow-cooling torsion-angle simulated annealing (started from
a very high temperature) drastically improved this initial
model to the point of allowing phasing through cycles of
model building and refinement to be initiated. The structure of
the protein is presented with emphasis on the presence of a
C(alpha)-modified proline at its active site, which was modelled as an
alpha-hydroxy-L-proline.
33. Kokkinidis, M., Glykos, N.M. & Fadouloglou*, V.E. (2012), "Protein Flexibility and Enzymatic Catalysis", Adv. Protein Chem. Struct. Biol., 87, 181-218.
Electronic reprint (562 KBytes) : Local copy, or directly from Adv. Protein Chem. Struct. Biol., Copyright © Elsevier.

 The dynamic nature of protein structures has been recognized, established,
and accepted as an intrinsic fundamental property with major
consequences to their function. Nowadays, proteins are considered as
networks of continuous motions, which reflect local flexibility and a propensity
for global structural plasticity. Proteinâprotein and proteinâsmall
ligand interactions, signal transduction and assembly of macromolecular
machines, allosteric regulation and thermal enzymatic adaptation are
processes which require structural flexibility. In general, enzymes represent
an attractive class among proteins in the study of protein flexibility
and they can be used as model systems for understanding the implications
of protein fluctuations to biological function. Flexibility of the active site is
considered as a requirement for reduction of free energy barrier and
acceleration of the enzymatic reaction while there is growing evidence
which concerns the connection between flexibility and substrate turnover
rate. Moreover, the role of conformational flexibility has been well established
in connection with the accessibility of the active site, the binding of
substrates and ligands, and release of products, stabilization and trapping
of intermediates, orientation of the substrate into the binding cleft, adjustment
of the reaction environment, etc.
The dynamic nature of protein structures has been recognized, established,
and accepted as an intrinsic fundamental property with major
consequences to their function. Nowadays, proteins are considered as
networks of continuous motions, which reflect local flexibility and a propensity
for global structural plasticity. Proteinâprotein and proteinâsmall
ligand interactions, signal transduction and assembly of macromolecular
machines, allosteric regulation and thermal enzymatic adaptation are
processes which require structural flexibility. In general, enzymes represent
an attractive class among proteins in the study of protein flexibility
and they can be used as model systems for understanding the implications
of protein fluctuations to biological function. Flexibility of the active site is
considered as a requirement for reduction of free energy barrier and
acceleration of the enzymatic reaction while there is growing evidence
which concerns the connection between flexibility and substrate turnover
rate. Moreover, the role of conformational flexibility has been well established
in connection with the accessibility of the active site, the binding of
substrates and ligands, and release of products, stabilization and trapping
of intermediates, orientation of the substrate into the binding cleft, adjustment
of the reaction environment, etc.
32. Georgoulia, P.S. & Glykos*, N.M. (2011), "Using J-coupling constants for force field validation: Application to hepta-alanine", J. Phys. Chem. B, 115, 15221â15227.
Electronic reprint (3.6 MBytes) : Local copy, or directly from J. Phys. Chem. B, Copyright © American Chemical Society. See also the Supporting Information.

 A computational solution to the protein folding problem is the holy grail of
biomolecular simulation and of the corresponding force fields. The
complexity of the systems used for folding simulations precludes a direct
feedback between the simulations and the force fields, thus necessitating
the study of simpler systems with sufficient experimental data to allow
force field optimization and validation. Recent studies on short polyalanine
peptides of increasing length (up to penta-alanine) indicated the presence
of a systematic deviation between the experimental (NMR-derived) J-couplings
and the great majority of biomolecular force fields, with the Ï2 values for
even the best-performing force fields being in the 1.4â1.8 range. Here we
show that by increasing the number of residues to seven and by achieving
convergence through an increase of the simulation time to 2 ÎŒs, we can
identify one force field (the AMBER99SB force field, out of the three force
fields studied) which when compared with the experimental J-coupling data
(and for a specific set of Karplus-equation parameters and estimated
J-coupling errors previously used in the literature) gave a value of
Ï2=0.99, indicating that full statistical consistency between experiment and
simulation is feasible. However, and as a detailed analysis of the effects
of estimated errors shows, the Ï2 values may be unsuitable as indicators of
the goodness-of-fit of the various biomolecular force fields.
A computational solution to the protein folding problem is the holy grail of
biomolecular simulation and of the corresponding force fields. The
complexity of the systems used for folding simulations precludes a direct
feedback between the simulations and the force fields, thus necessitating
the study of simpler systems with sufficient experimental data to allow
force field optimization and validation. Recent studies on short polyalanine
peptides of increasing length (up to penta-alanine) indicated the presence
of a systematic deviation between the experimental (NMR-derived) J-couplings
and the great majority of biomolecular force fields, with the Ï2 values for
even the best-performing force fields being in the 1.4â1.8 range. Here we
show that by increasing the number of residues to seven and by achieving
convergence through an increase of the simulation time to 2 ÎŒs, we can
identify one force field (the AMBER99SB force field, out of the three force
fields studied) which when compared with the experimental J-coupling data
(and for a specific set of Karplus-equation parameters and estimated
J-coupling errors previously used in the literature) gave a value of
Ï2=0.99, indicating that full statistical consistency between experiment and
simulation is feasible. However, and as a detailed analysis of the effects
of estimated errors shows, the Ï2 values may be unsuitable as indicators of
the goodness-of-fit of the various biomolecular force fields.
31. Patapati, K.K. & Glykos*, N.M. (2011), "Three Force Fields' Views of the 310 Helix", Biophys. J., 101, 1766-1771.
Electronic reprint (805 KBytes) : Local copy, or directly from Biophys. J., 101, 1766-1771. Copyright © 2011 Biophysical Society.

 Slowly but steadily bibliographic evidence is accumulating that the apparent
convergence of the various biomolecular force fields as evidenced from
simulations of proteins in the folded state does not hold true for folding
simulations. Here we add one more example to the growing list of peptides
and proteins for which different force fields show irreconcilable
differences in their folding predictions, even at such a fundamental level
as that of a peptide's secondary structure. We show that for an undecamer
peptide that is known from two independent NMR structure determinations to
have a mainly 310-helical structure in solution, three mainstream
biomolecular force fields give completely disparate predictions: The CHARMM
force field (with the CMAP correction) predicts an outstandingly stable
α-helical structure, in disagreement not only with the experimental
structures, but also with experimental evidence obtained from circular
dichroism. OPLS-AA shows an almost totally disordered peptide with the most
frequently observed folded conformation corresponding to a β-hairpin-like
structure, again in disagreement with all available experimental evidence.
Only the AMBER99SB force field appears to qualitatively agree with not only
the general structural characteristics of the peptide (on the account of
both NMR- and CD-based experiments), but to also correctly predict some of
the experimentally observed interactions at the level of side chains.
Possible interpretations of these findings are discussed.
Slowly but steadily bibliographic evidence is accumulating that the apparent
convergence of the various biomolecular force fields as evidenced from
simulations of proteins in the folded state does not hold true for folding
simulations. Here we add one more example to the growing list of peptides
and proteins for which different force fields show irreconcilable
differences in their folding predictions, even at such a fundamental level
as that of a peptide's secondary structure. We show that for an undecamer
peptide that is known from two independent NMR structure determinations to
have a mainly 310-helical structure in solution, three mainstream
biomolecular force fields give completely disparate predictions: The CHARMM
force field (with the CMAP correction) predicts an outstandingly stable
α-helical structure, in disagreement not only with the experimental
structures, but also with experimental evidence obtained from circular
dichroism. OPLS-AA shows an almost totally disordered peptide with the most
frequently observed folded conformation corresponding to a β-hairpin-like
structure, again in disagreement with all available experimental evidence.
Only the AMBER99SB force field appears to qualitatively agree with not only
the general structural characteristics of the peptide (on the account of
both NMR- and CD-based experiments), but to also correctly predict some of
the experimentally observed interactions at the level of side chains.
Possible interpretations of these findings are discussed.
30. Glykos*, N.M. (2011), "On the application of structure-specific bulk-solvent models", Acta Crystallogr., D67, 739-741.
Electronic reprint (982 KBytes) : Local copy, or directly from Acta Cryst. D67, 739-741, Copyright © International Union of Crystallography.

 It is often discussed, mainly in connection with the rather high
macromolecular R factors, that the treatment of bulk solvent in
macromolecular refinement may lack the detail needed for modelling the
solvent environment of molecules as complex as proteins and nucleic acids.
This line of thought directly leads to the hypothesis that improvements in
the modelling of the bulk solvent may substantially improve the agreement
between the experimental data and the crystallographic models. Here, part of
this hypothesis is being tested through the construction, via
molecular-dynamics simulations, of a highly detailed, physics-based,
structure-specific and crystallographic data-agnostic model of the bulk
solvent of a known crystal structure. The water-distribution map obtained
from the simulation is converted (after imposing space-group symmetry) to a
constant (but scalable) partial structure factor which is then added in a
re-refinement of the crystal structure. Compared with the simple
Babinet-based correction, a reduction of the totally cross-validated free R
value by 0.3% is observed. The implications and possible interpretations of
these results are discussed.
It is often discussed, mainly in connection with the rather high
macromolecular R factors, that the treatment of bulk solvent in
macromolecular refinement may lack the detail needed for modelling the
solvent environment of molecules as complex as proteins and nucleic acids.
This line of thought directly leads to the hypothesis that improvements in
the modelling of the bulk solvent may substantially improve the agreement
between the experimental data and the crystallographic models. Here, part of
this hypothesis is being tested through the construction, via
molecular-dynamics simulations, of a highly detailed, physics-based,
structure-specific and crystallographic data-agnostic model of the bulk
solvent of a known crystal structure. The water-distribution map obtained
from the simulation is converted (after imposing space-group symmetry) to a
constant (but scalable) partial structure factor which is then added in a
re-refinement of the crystal structure. Compared with the simple
Babinet-based correction, a reduction of the totally cross-validated free R
value by 0.3% is observed. The implications and possible interpretations of
these results are discussed.
29. Glykos*, N.M. (2011), "The 11th Misconception?", A letter to the Editor, CBE Life Sci Educ, 10(1), 1-2.
Electronic reprint (123 KBytes) : Local copy, or directly from CBE Life Sci Educ, 10(1), 1-2, Copyright © Glykos under the Creative Commons Attribution License.

 Biological sequences are an abstraction of an abstraction: In the first
level, we substituted the complexity of a proper three-dimensional entity
(like an amino acid residue) with a two dimensional chemical formula
describing only composition and covalent bonding. In the second abstraction
layer, we substituted these chemical formulas with single alphabet letters.
And then we forgot about it, and started behaving as if sequences do exist,
as if this artificial one-dimensionality is real. Sequence usage became so
widespread, that not only we forgot that these one-dimensional strings of
letters do not (and never did) exist, but we have started using them for
dealing with problems (like protein folding) that by their nature defy this
whole 'sequence' abstraction. Maybe, just maybe, we have had more than
enough of 'sequences' ?
Biological sequences are an abstraction of an abstraction: In the first
level, we substituted the complexity of a proper three-dimensional entity
(like an amino acid residue) with a two dimensional chemical formula
describing only composition and covalent bonding. In the second abstraction
layer, we substituted these chemical formulas with single alphabet letters.
And then we forgot about it, and started behaving as if sequences do exist,
as if this artificial one-dimensionality is real. Sequence usage became so
widespread, that not only we forgot that these one-dimensional strings of
letters do not (and never did) exist, but we have started using them for
dealing with problems (like protein folding) that by their nature defy this
whole 'sequence' abstraction. Maybe, just maybe, we have had more than
enough of 'sequences' ?
28. Patapati, K.K. & Glykos*, N.M. (2010), "Order through Disorder: Hyper-Mobile C-Terminal Residues Stabilize the Folded State of a Helical Peptide. A Molecular Dynamics Study", PLoS ONE, 5, e15290.
Electronic reprint (607 KBytes) : Local copy, or directly from PLos ONE, 5, e15290, Copyright © Patapati & Glykos under the Creative Commons Attribution License, See also the Supporting Information.

 Conventional wisdom has it that the presence of disordered regions in the
three-dimensional structures of polypeptides not only does not contribute
significantly to the thermodynamic stability of their folded state, but, on
the contrary, that the presence of disorder leads to a decrease of the
corresponding proteins' stability. We have performed extensive 3.4 ÎŒs long
folding simulations (in explicit solvent and with full electrostatics) of an
undecamer peptide of experimentally known helical structure, both with and
without its disordered (four residue long) C-terminal tail. Our simulations
clearly indicate that the presence of the apparently disordered (in
structural terms) C-terminal tail, increases the thermodynamic stability of
the peptide's folded (helical) state. These results show that at least for
the case of relatively short peptides, the interplay between thermodynamic
stability and the apparent structural stability can be rather subtle, with
even disordered regions contributing significantly to the stability of the
folded state. Our results have clear implications for the understanding of
peptide energetics and the design of foldable peptides.
Conventional wisdom has it that the presence of disordered regions in the
three-dimensional structures of polypeptides not only does not contribute
significantly to the thermodynamic stability of their folded state, but, on
the contrary, that the presence of disorder leads to a decrease of the
corresponding proteins' stability. We have performed extensive 3.4 ÎŒs long
folding simulations (in explicit solvent and with full electrostatics) of an
undecamer peptide of experimentally known helical structure, both with and
without its disordered (four residue long) C-terminal tail. Our simulations
clearly indicate that the presence of the apparently disordered (in
structural terms) C-terminal tail, increases the thermodynamic stability of
the peptide's folded (helical) state. These results show that at least for
the case of relatively short peptides, the interplay between thermodynamic
stability and the apparent structural stability can be rather subtle, with
even disordered regions contributing significantly to the stability of the
folded state. Our results have clear implications for the understanding of
peptide energetics and the design of foldable peptides.
27. Fadouloglou, V.E., Stavrakoudis, S., Bouriotis, V., Kokkinidis, M., & Glykos*, N.M. (2009), "Molecular Dynamics Simulations of BcZBP, A Deacetylase from Bacillus cereus: Active Site Loops Determine Substrate Accessibility and Specificity", J. Chem. Theory Comput., 5, 3299-3311.
Electronic reprint (7.3 MBytes) : Local copy, or directly from J. Chem. Theory Comput., 5, 3299-3311, Copyright © American Chemical Society.

 BcZBP is an LmbE-like, homohexameric, zinc-dependent deacetylase from the
opportunistic pathogen Bacillus cereus with three, thus far uncharacterized,
homologues in B. anthracis. Although its specific substrate is still
unknown, the enzyme has been shown to preferentially deacetylate
N-acetylglucosamine and diacetylchitobiose via an active site based on a
zinc-binding motif of the type HXDDXnH. In the crystal structure, the active
site is located at a deep and partially blocked cleft formed at the
interface between monomers related by the molecular 3-fold axis, although
the major, in structural terms, building block of the enzyme is not the
trimer, but the intertwined dimer. Here, we report results from a 50 ns
molecular dynamics simulation of BcZBP in explicit solvent with full
electrostatics and show that (i) the view of the intertwined dimer as the
major structural and functional building block of this class of hexameric
enzymes is possibly an oversimplification of the rather complex dynamics
observed in the simulation, (ii) the most mobile (with respect to their
atomic fluctuations) parts of the structure coincide with three surface
loops surrounding the active site, and (iii) these mobile loops define the
active site's accessibility, and may be implicated in the determination of
the enzyme's specificity.
BcZBP is an LmbE-like, homohexameric, zinc-dependent deacetylase from the
opportunistic pathogen Bacillus cereus with three, thus far uncharacterized,
homologues in B. anthracis. Although its specific substrate is still
unknown, the enzyme has been shown to preferentially deacetylate
N-acetylglucosamine and diacetylchitobiose via an active site based on a
zinc-binding motif of the type HXDDXnH. In the crystal structure, the active
site is located at a deep and partially blocked cleft formed at the
interface between monomers related by the molecular 3-fold axis, although
the major, in structural terms, building block of the enzyme is not the
trimer, but the intertwined dimer. Here, we report results from a 50 ns
molecular dynamics simulation of BcZBP in explicit solvent with full
electrostatics and show that (i) the view of the intertwined dimer as the
major structural and functional building block of this class of hexameric
enzymes is possibly an oversimplification of the rather complex dynamics
observed in the simulation, (ii) the most mobile (with respect to their
atomic fluctuations) parts of the structure coincide with three surface
loops surrounding the active site, and (iii) these mobile loops define the
active site's accessibility, and may be implicated in the determination of
the enzyme's specificity.
26. Fadouloglou, V.E., Bastaki, M.N., Ashcroft, A.E., Phillips, S.E.V., Panopoulos, N.J., Glykos*, N.M., & Kokkinidis*, M. (2009), "On the quaternary association of the type III secretion system HrcQB-C protein: Experimental evidence differentiates among the various oligomerization models", J. Struct. Biol., 166, 214-225.
Electronic reprint (3.7 MBytes) : Local copy, or directly from J. Struct. Biol., 166, 214-225, Copyright © Elsevier B.V.

 The HrcQB protein from the plant pathogen Pseudomonas syringae is a core
component of the bacterial type III secretion apparatus. The core consists
of nine proteins widely conserved among animal and plant pathogens which
also share sequence and structural similarities with proteins from the
bacterial flagellum. Previous studies of the carboxy-terminal domain of
HrcQB (HrcQB-C) and its flagellar homologue, FliN-C, have revealed
extensive sequence and structural homologies, similar subcellular
localization, and participation in analogous protein-protein interaction
networks. It is not clear however whether the similarities between the two
proteins extend to the level of quaternary association which is essential
for the formation of higher-order structures within the TTSS. Even though
the crystal structure of the FliN is a dimer, more detailed studies support
a tetrameric donut-like association. However, both models, dimer and
donut-like tetramer, are quite different from the crystallographic elongated
dimer of dimers of the HrcQB-C. To resolve this discrepancy we performed a
multidisciplinary investigation of the quaternary association of the
HrcQB-C, including mass-spectrometry, electrophoresis in non-reductive
conditions, gel filtration, glutaraldehyde cross-linking and small angle
X-ray scattering. Our experiments indicate that stable tetramers of
elongated shape are assembled in solution, in agreement with the results of
crystallographic studies. Circular dichroism data are consistent with a
dimer-dimer interface analogous to the one established in the crystal
structure. Finally, molecular dynamics simulations reveal the relative
orientation of the dimers forming the tetramers and the possible differences
from that of the crystal structure.
The HrcQB protein from the plant pathogen Pseudomonas syringae is a core
component of the bacterial type III secretion apparatus. The core consists
of nine proteins widely conserved among animal and plant pathogens which
also share sequence and structural similarities with proteins from the
bacterial flagellum. Previous studies of the carboxy-terminal domain of
HrcQB (HrcQB-C) and its flagellar homologue, FliN-C, have revealed
extensive sequence and structural homologies, similar subcellular
localization, and participation in analogous protein-protein interaction
networks. It is not clear however whether the similarities between the two
proteins extend to the level of quaternary association which is essential
for the formation of higher-order structures within the TTSS. Even though
the crystal structure of the FliN is a dimer, more detailed studies support
a tetrameric donut-like association. However, both models, dimer and
donut-like tetramer, are quite different from the crystallographic elongated
dimer of dimers of the HrcQB-C. To resolve this discrepancy we performed a
multidisciplinary investigation of the quaternary association of the
HrcQB-C, including mass-spectrometry, electrophoresis in non-reductive
conditions, gel filtration, glutaraldehyde cross-linking and small angle
X-ray scattering. Our experiments indicate that stable tetramers of
elongated shape are assembled in solution, in agreement with the results of
crystallographic studies. Circular dichroism data are consistent with a
dimer-dimer interface analogous to the one established in the crystal
structure. Finally, molecular dynamics simulations reveal the relative
orientation of the dimers forming the tetramers and the possible differences
from that of the crystal structure.
25. Fadouloglou, V.E., Kokkinidis, M. & Glykos*, N.M. (2008), "Determination of protein oligomerization state: Two approaches based on glutaraldehyde crosslinking", Anal. Biochem., 373, 404-406.
Electronic reprint (605 KBytes) : Local copy, or directly from Anal. Biochem., 373, 404-406, Copyright © Elsevier B.V.

 Many biochemical and biophysical methods can be used to
characterize the oligomerization state of proteins. One of the most widely
used is glutaraldehyde crosslinking, mainly because of the minimum equipment
and reagents required. However, the crosslinking procedures currently in use
are impaired by the low specificity of the reagent, which can chemically
bond any two amino groups that are close in space. Thus, extensive and
time-consuming investigation of the reaction conditions is usually required.
Here we describe two approaches based on glutaraldehyde that readily give
reliable results.
Many biochemical and biophysical methods can be used to
characterize the oligomerization state of proteins. One of the most widely
used is glutaraldehyde crosslinking, mainly because of the minimum equipment
and reagents required. However, the crosslinking procedures currently in use
are impaired by the low specificity of the reagent, which can chemically
bond any two amino groups that are close in space. Thus, extensive and
time-consuming investigation of the reaction conditions is usually required.
Here we describe two approaches based on glutaraldehyde that readily give
reliable results.
24. Mizas, Ch., Sirakoulis, G.Ch., Mardiris, V., Karafyllidis, I., Glykos, N.M. & Sandaltzopoulos*, R. (2008), "Reconstruction of DNA sequences using genetic algorithms and cellular automata: Towards mutation prediction ?", BioSystems, 92, 61-68.
Electronic reprint (574 KBytes) : Local copy, or directly from BioSystems, 92, 61-68, Copyright © Elsevier B.V.

 Change of DNA sequence that fuels evolution is, to a certain
extent, a deterministic process because mutagenesis does not occur in an
absolutely random manner. So far, it has not been possible to decipher the
rules that govern DNA sequence evolution due to the extreme complexity of
the entire process. In our attempt to approach this issue we focus solely on
the mechanisms of mutagenesis and deliberately disregard the role of natural
selection. Hence, in this analysis, evolution refers to the accumulation of
genetic alterations that originate from mutations and are transmitted
through generations without being subjected to natural selection. We have
developed a software tool that allows modelling of a DNA sequence as a
one-dimensional cellular automaton (CA) with four states per cell which
correspond to the four DNA bases, i.e. A, C, T and G. The four states are
represented by numbers of the quaternary number system. Moreover, we have
developed genetic algorithms (GAs) in order to determine the rules of CA
evolution that simulate the DNA evolution process. Linear evolution rules
were considered and square matrices were used to represent them. If DNA
sequences of different evolution steps are available, our approach allows
the determination of the underlying evolution rule(s). Conversely, once the
evolution rules are deciphered, our tool may reconstruct the DNA sequence in
any previous evolution step for which the exact sequence information was
unknown. The developed tool may be used to test various parameters that
could influence evolution. We describe a paradigm relying on the assumption
that mutagenesis is governed by a near-neighbour-dependent mechanism. Based
on the satisfactory performance of our system in the deliberately
simplified example, we propose that our approach could offer a starting
point for future attempts to understand the mechanisms that govern
evolution. The developed software is open-source and has a user-friendly
graphical input interface.
Change of DNA sequence that fuels evolution is, to a certain
extent, a deterministic process because mutagenesis does not occur in an
absolutely random manner. So far, it has not been possible to decipher the
rules that govern DNA sequence evolution due to the extreme complexity of
the entire process. In our attempt to approach this issue we focus solely on
the mechanisms of mutagenesis and deliberately disregard the role of natural
selection. Hence, in this analysis, evolution refers to the accumulation of
genetic alterations that originate from mutations and are transmitted
through generations without being subjected to natural selection. We have
developed a software tool that allows modelling of a DNA sequence as a
one-dimensional cellular automaton (CA) with four states per cell which
correspond to the four DNA bases, i.e. A, C, T and G. The four states are
represented by numbers of the quaternary number system. Moreover, we have
developed genetic algorithms (GAs) in order to determine the rules of CA
evolution that simulate the DNA evolution process. Linear evolution rules
were considered and square matrices were used to represent them. If DNA
sequences of different evolution steps are available, our approach allows
the determination of the underlying evolution rule(s). Conversely, once the
evolution rules are deciphered, our tool may reconstruct the DNA sequence in
any previous evolution step for which the exact sequence information was
unknown. The developed tool may be used to test various parameters that
could influence evolution. We describe a paradigm relying on the assumption
that mutagenesis is governed by a near-neighbour-dependent mechanism. Based
on the satisfactory performance of our system in the deliberately
simplified example, we propose that our approach could offer a starting
point for future attempts to understand the mechanisms that govern
evolution. The developed software is open-source and has a user-friendly
graphical input interface.
23. Fadouloglou, V.E., Deli, A., Glykos, N.M., Psylinakis, E., Bouriotis, V. & Kokkinidis*, M. (2007), "Crystal structure of the BcZBP, a zinc-binding protein from Bacillus cereus. Functional insights from structural data", FEBS J., 274, 3044-3054.
Electronic reprint (1.2 MBytes) : Local copy, or directly from FEBS J., 274, 3044-3054, Copyright © Blackwell Publishing, Inc.

 Bacillus cereus is an opportunistic pathogenic bacterium closely related to
B. anthracis, the causative agent of anthrax in mammals. A
significant portion of the B. cereus chromosomal genes are common to
B. anthracis, including genes which in B. anthracis code for putative virulence
and surface proteins. B. cereus thus provides a convenient model organism
for studying proteins potentially associated with the pathogenicity of the
highly infectious B. anthracis. The zinc-binding protein of B. cereus,
BcZBP, is encoded from the bc1534 gene which has three homologues to
B. anthracis. The protein exhibits deacetylase activity with the N-acetyl
moiety of the N-acetylglucosamine and the diacetylchitobiose and
triacetylchitotriose. However, neither the specific substrate of the BcZBP
nor the biochemical pathway have been conclusively identified. Here, we
present the crystal structure of BcZBP at 1.8 A resolution. The
N-terminal part of the 234 amino acid protein adopts a Rossmann fold whereas
the C-terminal part consists of two beta strands and two alpha helices. In the
crystal, the protein forms a compact hexamer, in agreement with solution
data. A zinc binding site and a potential active site have been identified
in each monomer. These sites have extensive similarities to those found in
two known zinc-dependent hydrolases with deacetylase activity, MshB and
LpxC, despite a low degree of amino acid sequence identity. The functional
implications and a possible catalytic mechanism are discussed.
(2ixd.pdb).
Bacillus cereus is an opportunistic pathogenic bacterium closely related to
B. anthracis, the causative agent of anthrax in mammals. A
significant portion of the B. cereus chromosomal genes are common to
B. anthracis, including genes which in B. anthracis code for putative virulence
and surface proteins. B. cereus thus provides a convenient model organism
for studying proteins potentially associated with the pathogenicity of the
highly infectious B. anthracis. The zinc-binding protein of B. cereus,
BcZBP, is encoded from the bc1534 gene which has three homologues to
B. anthracis. The protein exhibits deacetylase activity with the N-acetyl
moiety of the N-acetylglucosamine and the diacetylchitobiose and
triacetylchitotriose. However, neither the specific substrate of the BcZBP
nor the biochemical pathway have been conclusively identified. Here, we
present the crystal structure of BcZBP at 1.8 A resolution. The
N-terminal part of the 234 amino acid protein adopts a Rossmann fold whereas
the C-terminal part consists of two beta strands and two alpha helices. In the
crystal, the protein forms a compact hexamer, in agreement with solution
data. A zinc binding site and a potential active site have been identified
in each monomer. These sites have extensive similarities to those found in
two known zinc-dependent hydrolases with deacetylase activity, MshB and
LpxC, despite a low degree of amino acid sequence identity. The functional
implications and a possible catalytic mechanism are discussed.
(2ixd.pdb).
22. Glykos*, N.M. (2007), "On the application of molecular-dynamics simulations to validate thermal parameters and to optimize TLS-group selection for macromolecular refinement", Acta Crystallogr., D63, 705-713.
Electronic reprint (1.4 MBytes) : Local copy, or directly from Acta Cryst. D63, 705-713, Copyright © International Union of Crystallography.

 Comparison of crystallographically determined and molecular dynamics
simulation-derived parameters for a small (26 kDa) homotetrameric
four-alpha-helical bundle protein revealed an unexpected pattern of similarities
and differences between experiment and simulation. On one hand, the protein
structure per se is exceptionally well preserved during the simulations,
with a root-mean-square deviation between the Ca atoms of the crystal
structure and the simulation-derived average structures of only 0.58 Angstrom,
which is not very different from the expected coordinate error of the
experimentally determined structure. On the other hand, comparison of the
temperature factors showed a large discrepancy, with the experimental B-factors
being approximately three times higher than the simulation-derived B-factors.
Closer examination of this discrepancy appears to validate the
molecular dynamics prediction and to implicate as its source static disorder
at the crystalline state, as indicated by the strong diffuse scattering and
pronounced anisotropy of the diffraction pattern of the protein crystals.
A posteriori re-refinement of the structure using a new TLS parameterization
scheme based on the results obtained from the simulations led to a further
reduction of the R factor and the free R value by 0.4% and 0.8%,
respectively, indicating that molecular-dynamics simulations have matured to
the point that they can be used to aid the selection of TLS groups for
macromolecular refinement.
Comparison of crystallographically determined and molecular dynamics
simulation-derived parameters for a small (26 kDa) homotetrameric
four-alpha-helical bundle protein revealed an unexpected pattern of similarities
and differences between experiment and simulation. On one hand, the protein
structure per se is exceptionally well preserved during the simulations,
with a root-mean-square deviation between the Ca atoms of the crystal
structure and the simulation-derived average structures of only 0.58 Angstrom,
which is not very different from the expected coordinate error of the
experimentally determined structure. On the other hand, comparison of the
temperature factors showed a large discrepancy, with the experimental B-factors
being approximately three times higher than the simulation-derived B-factors.
Closer examination of this discrepancy appears to validate the
molecular dynamics prediction and to implicate as its source static disorder
at the crystalline state, as indicated by the strong diffuse scattering and
pronounced anisotropy of the diffraction pattern of the protein crystals.
A posteriori re-refinement of the structure using a new TLS parameterization
scheme based on the results obtained from the simulations led to a further
reduction of the R factor and the free R value by 0.4% and 0.8%,
respectively, indicating that molecular-dynamics simulations have matured to
the point that they can be used to aid the selection of TLS groups for
macromolecular refinement.
21. Glykos*, N.M. (2006), "Carma: a molecular dynamics analysis program", J. Comput. Chem., 27, 1765-1768.
Electronic reprint (284 KBytes) : Local copy, or directly from Wiley InterScience, Copyright © Wiley InterScience.
 A computer program has been developed to aid the analysis of
molecular dynamics trajectories. The program is tuned for macromolecular
large-scale problems and supports features such as removal of global
translations-rotations of the solute, calculation of average distance maps
and their corresponding standard deviations, calculation of the
variance-covariance and cross-correlation matrices, and principal component
analysis of trajectories with the added ability to create artificial
trajectories based on selected eigenvectors. Limited graphics (trajectory
viewing) capabilities are also available.
A computer program has been developed to aid the analysis of
molecular dynamics trajectories. The program is tuned for macromolecular
large-scale problems and supports features such as removal of global
translations-rotations of the solute, calculation of average distance maps
and their corresponding standard deviations, calculation of the
variance-covariance and cross-correlation matrices, and principal component
analysis of trajectories with the added ability to create artificial
trajectories based on selected eigenvectors. Limited graphics (trajectory
viewing) capabilities are also available.
20. Glykos, N.M., Papanikolau, Y., Vlassi, M., Kotsifaki, D., Cesareni G. & Kokkinidis*, M. (2006), "Loopless Rop: Structure and Dynamics of an Engineered Homotetrameric Variant of the Repressor of Primer Protein", Biochemistry, 45, 10905-10919.
Electronic reprint (2.0 MBytes) : Local copy, or directly from the American Chemical Society, Copyright © American Chemical Society.
 The repressor of primer (Rop) protein has become a steady source
of surprises concerning the relationship between the sequences and the
structures of several of its mutants and variants. Here we add another piece
to the puzzle of Rop by showing that an engineered deletion mutant of the
protein (corresponding to a deletion of residues 30-34 of the wild-type
protein and designed to restore the heptad periodicity at the turn region)
results in a complete reorganization of the bundle which is converted from a
homodimer to a homotetramer. In contrast (and as previously shown), a
two-residue insertion, which also restores the heptad periodicity, is
essentially identical with wild-type Rop. The new deletion mutant structure
is a canonical, left-handed, all-antiparallel bundle with a completely
different hydrophobic core and distinct surface properties. The structure
agrees and qualitatively explains the results from functional,
thermodynamic, and kinetic studies which indicated that this deletion mutant
is a biologically inactive hyperstable homotetramer. Additional insight into
the stability and dynamics of the mutant structure has been obtained from
extensive molecular dynamics simulations in explicit water and with full
treatment of electrostatics.
(1qx8.pdb).
The repressor of primer (Rop) protein has become a steady source
of surprises concerning the relationship between the sequences and the
structures of several of its mutants and variants. Here we add another piece
to the puzzle of Rop by showing that an engineered deletion mutant of the
protein (corresponding to a deletion of residues 30-34 of the wild-type
protein and designed to restore the heptad periodicity at the turn region)
results in a complete reorganization of the bundle which is converted from a
homodimer to a homotetramer. In contrast (and as previously shown), a
two-residue insertion, which also restores the heptad periodicity, is
essentially identical with wild-type Rop. The new deletion mutant structure
is a canonical, left-handed, all-antiparallel bundle with a completely
different hydrophobic core and distinct surface properties. The structure
agrees and qualitatively explains the results from functional,
thermodynamic, and kinetic studies which indicated that this deletion mutant
is a biologically inactive hyperstable homotetramer. Additional insight into
the stability and dynamics of the mutant structure has been obtained from
extensive molecular dynamics simulations in explicit water and with full
treatment of electrostatics.
(1qx8.pdb).
19. Glykos*, N.M. (2005), "Qs v.1.3: a parallel version of Queen of Spades", J. Appl. Crystallogr., 38, 574-575.
Electronic reprint (65 KBytes) : Local copy, or directly from J. Appl. Crystallogr. 38, 574-575, Copyright © International Union of Crystallography.
 The program Queen of Spades encodes an algorithm for a stochastic multidimensional approach to
molecular replacement. The program has been shown to be capable of successfully
locating solutions even in cases as complex as a 23-dimensional, 4-body
problem. Recently, we extended our approach
to tackle the full molecular replacement problem by allowing the possibility
of using many different search models simultaneously, and showed that we
could successfully locate solutions in the case of a 17-dimensional problem
involving one DNA and two (different) protein search models.
This multimodel, multidimensional approach does have
its cost: with a few thousand unique reflections in a high symmetry space
group and with more than two search models, a typical Qs run would
take well over two to three weeks of CPU time on the fastest personal
workstations. The way forward for such computationally intensive
calculations is of course parallelisation. Here, I report the availability of
a parallel version of Qs which is based on the Message Passing
Interface (MPI) paradigm.
The program Queen of Spades encodes an algorithm for a stochastic multidimensional approach to
molecular replacement. The program has been shown to be capable of successfully
locating solutions even in cases as complex as a 23-dimensional, 4-body
problem. Recently, we extended our approach
to tackle the full molecular replacement problem by allowing the possibility
of using many different search models simultaneously, and showed that we
could successfully locate solutions in the case of a 17-dimensional problem
involving one DNA and two (different) protein search models.
This multimodel, multidimensional approach does have
its cost: with a few thousand unique reflections in a high symmetry space
group and with more than two search models, a typical Qs run would
take well over two to three weeks of CPU time on the fastest personal
workstations. The way forward for such computationally intensive
calculations is of course parallelisation. Here, I report the availability of
a parallel version of Qs which is based on the Message Passing
Interface (MPI) paradigm.
18. Glykos, N.M. & Kokkinidis*, M. (2004), "Structural Polymorphism of a Marginally Stable 4-alpha-Helical Bundle. Images of a Trapped Molten Globule ?", Proteins, 56, 420-425.
Electronic reprint (312 KBytes) : Local copy, or directly from Wiley InterScience, Copyright © Wiley InterScience.
 The Repressor of primer (Rop) protein is the paradigm of a canonical
(left-handed, all-anti-parallel) homodimeric 4-alpha-helical bundle. It
is known (through the analysis of an orthorhombic crystal form) that a
single alanine-to-proline amino-acid substitution at the turn region of Rop
(the A31P mutant) markedly changes the topology of the protein which is
converted to a right-handed, mixed parallel and antiparallel bundle. Here we
report the structure of this mutant in a second (monoclinic) crystal form
and show that although its topology remains unchanged, the differences
between the two A31P crystal structures are unexpectedly large, with a root
mean square deviation between equivalent Ca atoms of approximately
3A. Remarkably, a 3 ns molecular dynamics simulation of A31P which was
initiated from the orthorhombic form crystal structure, sampled an ensemble
of configurations rather similar to the structure seen in the monoclinic
form. This finding suggests that the observed crystal structures may
correspond to images of two conformers taken from a structurally
heterogeneous population of molecules at equilibrium. Comparison of the
A31P simulation with a 3 ns simulation of wild-type Rop indicated that the
mutant is an inherently highly flexible molecule, both with respect to the
relative placement of its helices and the malleability of its (loosely
packed) hydrophobic core. Based on these findings, we propose that the A31P
Rop mutant is an equilibrium molten globule and we attempt to interpret its
thermodynamic properties based on this assumption. Furthermore, we present
results from a 3 ns molecular dynamics simulation of a hypothetical
structure which was constructed by artificially mutating the alanine at
position 31 of the wild-type Rop structure to a proline. This hypothetical
A31P structure appears to be significantly more stable than the
experimentally determined one, leading us to propose that the observed A31P
structure may correspond to a kinetically trapped molten globule.
(1gmg.pdb).
The Repressor of primer (Rop) protein is the paradigm of a canonical
(left-handed, all-anti-parallel) homodimeric 4-alpha-helical bundle. It
is known (through the analysis of an orthorhombic crystal form) that a
single alanine-to-proline amino-acid substitution at the turn region of Rop
(the A31P mutant) markedly changes the topology of the protein which is
converted to a right-handed, mixed parallel and antiparallel bundle. Here we
report the structure of this mutant in a second (monoclinic) crystal form
and show that although its topology remains unchanged, the differences
between the two A31P crystal structures are unexpectedly large, with a root
mean square deviation between equivalent Ca atoms of approximately
3A. Remarkably, a 3 ns molecular dynamics simulation of A31P which was
initiated from the orthorhombic form crystal structure, sampled an ensemble
of configurations rather similar to the structure seen in the monoclinic
form. This finding suggests that the observed crystal structures may
correspond to images of two conformers taken from a structurally
heterogeneous population of molecules at equilibrium. Comparison of the
A31P simulation with a 3 ns simulation of wild-type Rop indicated that the
mutant is an inherently highly flexible molecule, both with respect to the
relative placement of its helices and the malleability of its (loosely
packed) hydrophobic core. Based on these findings, we propose that the A31P
Rop mutant is an equilibrium molten globule and we attempt to interpret its
thermodynamic properties based on this assumption. Furthermore, we present
results from a 3 ns molecular dynamics simulation of a hypothetical
structure which was constructed by artificially mutating the alanine at
position 31 of the wild-type Rop structure to a proline. This hypothetical
A31P structure appears to be significantly more stable than the
experimentally determined one, leading us to propose that the observed A31P
structure may correspond to a kinetically trapped molten globule.
(1gmg.pdb).
17. Papanikolau, Y., Kotsifaki, D., Fadouloglou, V.E., Gazi, A.D., Glykos, N.M., Cesareni G. & Kokkinidis*, M. (2004), "Ionic strength reducers: an efficient approach to protein purification and crystallization. Application to two Rop variants.", Acta Crystallogr., D60, 1334-1337.
Electronic reprint (539 KBytes) : Local copy, or directly from Acta Cryst. D60, 1334-1337, Copyright © International Union of Crystallography.
 Detailed knowledge of the influence of various parameters on
macromolecular solubility is essential for crystallization. The concept
of so-called ionic strength reducers provides insight into the changes
in solubility induced by organic solvents and hydrophilic polymers in
aqueous electrolytic solutions. A simple and efficient procedure is
presented which exploits the properties of ionic strength reducers in
the purification and crystallization of proteins. Using two designed
variants of the Rop protein as model systems, superior crystals have
been obtained compared with conventional techniques. This
procedure is particularly useful in cases where excessive nucleation
leads to the growth of a large number of tiny crystals that are useless
for crystallographic analysis.
Detailed knowledge of the influence of various parameters on
macromolecular solubility is essential for crystallization. The concept
of so-called ionic strength reducers provides insight into the changes
in solubility induced by organic solvents and hydrophilic polymers in
aqueous electrolytic solutions. A simple and efficient procedure is
presented which exploits the properties of ionic strength reducers in
the purification and crystallization of proteins. Using two designed
variants of the Rop protein as model systems, superior crystals have
been obtained compared with conventional techniques. This
procedure is particularly useful in cases where excessive nucleation
leads to the growth of a large number of tiny crystals that are useless
for crystallographic analysis.
16. Glykos*, N.M. & Kokkinidis, M. (2004), "Molecular Replacement with multiple different models", J. Appl. Crystallogr., 37, 159-161.
Electronic reprint (1.1 MBytes) : Local copy, or directly from J. Appl. Crystallogr. 37, 159-161, Copyright © International Union of Crystallography.
 Classical molecular replacement methods and the newer six-dimensional searches
treat molecular replacement as a succession of sub-problems of reduced dimensionality.
Due to their divide-and-conquer approach, these methods necessarily ignore (at
least during their early stages) the very knowledge that a target crystal structure may
comprise, for example, more than one copy of a search model, or, several models of
different types. We have previously described an algorithm for a stochastic multidimensional
molecular replacement search and showed that it can successfully locate
solutions even in cases as complex as a 23-dimensional, 4-body search. The original
description of the method only dealt with a special case of molecular replacement,
namely with the problem of placing n copies of only one search model in the
asymmetric unit of a target crystal structure. Here we present a natural generalisation of
this algorithm to deal with the full molecular replacement problem, that is, with the
problem of determining the orientations and positions of a total of n
copies of m different models which are assumed to be present in the asymmetric unit
of a target crystal structure. The generality of this approach is illustrated through its
successful application to a 17-dimensional, 3-model problem involving one DNA and
two protein molecules.
Classical molecular replacement methods and the newer six-dimensional searches
treat molecular replacement as a succession of sub-problems of reduced dimensionality.
Due to their divide-and-conquer approach, these methods necessarily ignore (at
least during their early stages) the very knowledge that a target crystal structure may
comprise, for example, more than one copy of a search model, or, several models of
different types. We have previously described an algorithm for a stochastic multidimensional
molecular replacement search and showed that it can successfully locate
solutions even in cases as complex as a 23-dimensional, 4-body search. The original
description of the method only dealt with a special case of molecular replacement,
namely with the problem of placing n copies of only one search model in the
asymmetric unit of a target crystal structure. Here we present a natural generalisation of
this algorithm to deal with the full molecular replacement problem, that is, with the
problem of determining the orientations and positions of a total of n
copies of m different models which are assumed to be present in the asymmetric unit
of a target crystal structure. The generality of this approach is illustrated through its
successful application to a 17-dimensional, 3-model problem involving one DNA and
two protein molecules.
15. Fadouloglou, V.E., Tampakaki, A.P., Glykos, N.M., Bastaki, N., Hadden, J.M., Phillips, S.E., Panopoulos, N.J. & Kokkinidis*, M. (2004), "Structure of HrcQb-C, a conserved component of the bacterial type III secretion systems", Proc. Natl. Acad. Sci. USA, 101, 70-75.
Electronic reprint (629 KBytes) : Local copy, or directly from Proc. Natl. Acad. Sci. USA, 101, 70-75, Copyright © National Academy of Sciences USA.
 Type III secretion systems enable plant and animal bacterial pathogens
to deliver virulence proteins into the cytosol of eukaryotic
host cells, causing a broad spectrum of diseases including
bacteremia, septicemia, typhoid fever, and bubonic plague in mammals,
and localized lesions, systemic wilting, and blights in plants. In
addition, type III secretion systems are also required for biogenesis
of the bacterial flagellum. The HrcQB protein, a component of the
secretion apparatus of Pseudomonas syringae with homologues in
all type III systems, has a variable N-terminal and a conserved
C-terminal domain (HrcQB-C). Here, we report the crystal structure
of HrcQB-C and show that this domain retains the ability of the
full-length protein to interact with other type III components. A 3D
analysis of sequence conservation patterns reveals two clusters of
residues potentially involved in proteinprotein interactions.
Based on the analogies between HrcQB and its flagellum homo-
logues, we propose that HrcQB-C participates in the formation of
a C-ring-like assembly.
(1o9y.pdb).
Type III secretion systems enable plant and animal bacterial pathogens
to deliver virulence proteins into the cytosol of eukaryotic
host cells, causing a broad spectrum of diseases including
bacteremia, septicemia, typhoid fever, and bubonic plague in mammals,
and localized lesions, systemic wilting, and blights in plants. In
addition, type III secretion systems are also required for biogenesis
of the bacterial flagellum. The HrcQB protein, a component of the
secretion apparatus of Pseudomonas syringae with homologues in
all type III systems, has a variable N-terminal and a conserved
C-terminal domain (HrcQB-C). Here, we report the crystal structure
of HrcQB-C and show that this domain retains the ability of the
full-length protein to interact with other type III components. A 3D
analysis of sequence conservation patterns reveals two clusters of
residues potentially involved in proteinprotein interactions.
Based on the analogies between HrcQB and its flagellum homo-
logues, we propose that HrcQB-C participates in the formation of
a C-ring-like assembly.
(1o9y.pdb).
14. Glykos*, N.M. & Kokkinidis, M. (2003), "Structure determination of a small protein through a 23-dimensional molecular replacement search", Acta Crystallogr., D59, 709-718.
Electronic reprint (3.8 MBytes) : Local copy, or directly from Acta Cryst. D59, 709-718, Copyright © International Union of Crystallography.
 The crystal structure of a 4-alpha-helical bundle protein has been
determined through the application of a 23-dimensional molecular replacement
search performed with a stochastic method. The search model for the
calculation was a 26 residue-long poly-alanine helix amounting to less than
13% of the total number of atoms in the asymmetric unit of the target
crystal structure. The crystal structure determination procedure is
presented in detail, with emphasis on the molecular replacement
calculations
(1gmg.pdb).
The crystal structure of a 4-alpha-helical bundle protein has been
determined through the application of a 23-dimensional molecular replacement
search performed with a stochastic method. The search model for the
calculation was a 26 residue-long poly-alanine helix amounting to less than
13% of the total number of atoms in the asymmetric unit of the target
crystal structure. The crystal structure determination procedure is
presented in detail, with emphasis on the molecular replacement
calculations
(1gmg.pdb).
13. Dennis, C.A., Glykos, N.M., Parsons, M.R. & Phillips*, S.E.V. (2002), "The structure of AhrC, the arginine repressor/activator protein from Bacillus subtilis", Acta Crystallogr., D58, 421-430.
Electronic reprint (1.6 MBytes) : Local copy, or directly from Acta Cryst. D58, 421-430, Copyright © International Union of Crystallography.

 In the gram positive bacterium Bacillus subtilis the concentration of the amino acid L-arginine is controlled
by the transcriptional regulator AhrC. The hexameric AhrC protein binds in an L-arginine-dependent manner to
pseudo-palindromic operators within the prometer regions of arginine biosynthesic and catabolic gene clusters.
AhrC binding results in the repression of transcription of biosynthetic genes and in the activation of transcription
of catabolic genes. We have determined the crystal structure of AhrC at 2.7A resulution. Each sununit of the protein
has two domains. The C-terminal domains are arranged with 32 point group symmetry and mediate the major inter-subunit
interactions. The N-terminal domains are located around this core, where they lie in weakly associated pairs but do not
obey strict symmetry. A structural comparison of AhrC with the arginine repressor from the thermophile
Bacillus stearothermophilus reveals close similarity in regions implicated in L-arginine binding and DNA recognition
but also some striking sequence differences, especially within the C-terminal oligomerisation domain, which may contribute
to the different thermostabilities of the proteins. Comparison of the crystal structure of AhrC with a 30A resolution
model obtained by combining X-ray structure factor amplitudes with phases derived from electron microscopic analyses of
AhrC crystals confirms the essential accuracy of the earlier model and suggests that such an approach may be more
widely useful for obtaining low resolution phase information
(1f9n.pdb).
In the gram positive bacterium Bacillus subtilis the concentration of the amino acid L-arginine is controlled
by the transcriptional regulator AhrC. The hexameric AhrC protein binds in an L-arginine-dependent manner to
pseudo-palindromic operators within the prometer regions of arginine biosynthesic and catabolic gene clusters.
AhrC binding results in the repression of transcription of biosynthetic genes and in the activation of transcription
of catabolic genes. We have determined the crystal structure of AhrC at 2.7A resulution. Each sununit of the protein
has two domains. The C-terminal domains are arranged with 32 point group symmetry and mediate the major inter-subunit
interactions. The N-terminal domains are located around this core, where they lie in weakly associated pairs but do not
obey strict symmetry. A structural comparison of AhrC with the arginine repressor from the thermophile
Bacillus stearothermophilus reveals close similarity in regions implicated in L-arginine binding and DNA recognition
but also some striking sequence differences, especially within the C-terminal oligomerisation domain, which may contribute
to the different thermostabilities of the proteins. Comparison of the crystal structure of AhrC with a 30A resolution
model obtained by combining X-ray structure factor amplitudes with phases derived from electron microscopic analyses of
AhrC crystals confirms the essential accuracy of the earlier model and suggests that such an approach may be more
widely useful for obtaining low resolution phase information
(1f9n.pdb).
12. Glykos*, N.M. & Kokkinidis, M. (2001), "Multidimensional Molecular Replacement", Acta Crystallogr., D57, 1462-1473.
Electronic reprint (1.9 MBytes) : Local copy, or directly from Acta Cryst. D57, 1462-1473, Copyright © International Union of Crystallography.
The ECM 20 (Krakow, 2001) presentation is also available for download (Powerpoint presentation, 1.6 MB).
 A method is described which attempts to simultaneously and independently
determine the positional and orientational parameters of all molecules
present in the asymmetric unit of a target crystal structure. This is
achieved through a reverse Monte Carlo optimisation of a suitable statistic
(like the R-factor or the linear correlation coefficient between the
observed and calculated amplitudes of the structure factors) in the
6n-dimensional space defined by the rotational and translational
parameters of the n search models. Results from the application of this
stochastic method ---obtained with a space group general computer program
which has been developed for this purpose--- indicate that with present-day
computing capabilities the method may successfully be applied to molecular
replacement problems for which the target crystal structure contains up to
three molecules per asymmetric unit. It is also shown that the method may be
useful in cases where the assumption of topological segregation of the self
and cross vectors in the Patterson function is violated (as may happen, for
example, in closely packed crystal structures).
A method is described which attempts to simultaneously and independently
determine the positional and orientational parameters of all molecules
present in the asymmetric unit of a target crystal structure. This is
achieved through a reverse Monte Carlo optimisation of a suitable statistic
(like the R-factor or the linear correlation coefficient between the
observed and calculated amplitudes of the structure factors) in the
6n-dimensional space defined by the rotational and translational
parameters of the n search models. Results from the application of this
stochastic method ---obtained with a space group general computer program
which has been developed for this purpose--- indicate that with present-day
computing capabilities the method may successfully be applied to molecular
replacement problems for which the target crystal structure contains up to
three molecules per asymmetric unit. It is also shown that the method may be
useful in cases where the assumption of topological segregation of the self
and cross vectors in the Patterson function is violated (as may happen, for
example, in closely packed crystal structures).
11. Fadouloglou, V.E., Glykos, N.M. & Kokkinidis*, M. (2001), "Side-chain conformations in 4-alpha-helical bundles", Protein Engng., 14, 321-328.
Electronic reprint (385 KBytes) : Local copy, or directly from Protein Engng. 14, 321-328, Copyright © Oxford University Press.

 The distribution of the chi1, chi2 dihedral angles in a data set consisting of
twelve unrelated 4-alpha-helical bundle proteins has been determined and
compared with that observed in globular proteins. The analysis suggests that
for this tertiary motif : (i) the side-chain conformations are limited to only
a subset of the conformations observed in globular proteins and are more
constrained that side chains in helical regions of globular proteins.
(ii) The side chains of Aspartic acid and Asparagine adopt occasionally a new,
topology-specific rotamer. (iii) The rotamer preferences of Tyrosine and
Isoleucine depend on whether they are located in the hydrophobic core of the
bundle or in a more exposed position. (iv) Naturally occuring mutations in
the hydrophobic core of 4-alpha-helical bundles follow a pattern that is
consistent with the notion that the mutated and wild-type residues have at
least one of their most highly populated rotamers in common. These findings
indicate a relationship between protein topology and preferred side-chains
conformations.
The distribution of the chi1, chi2 dihedral angles in a data set consisting of
twelve unrelated 4-alpha-helical bundle proteins has been determined and
compared with that observed in globular proteins. The analysis suggests that
for this tertiary motif : (i) the side-chain conformations are limited to only
a subset of the conformations observed in globular proteins and are more
constrained that side chains in helical regions of globular proteins.
(ii) The side chains of Aspartic acid and Asparagine adopt occasionally a new,
topology-specific rotamer. (iii) The rotamer preferences of Tyrosine and
Isoleucine depend on whether they are located in the hydrophobic core of the
bundle or in a more exposed position. (iv) Naturally occuring mutations in
the hydrophobic core of 4-alpha-helical bundles follow a pattern that is
consistent with the notion that the mutated and wild-type residues have at
least one of their most highly populated rotamers in common. These findings
indicate a relationship between protein topology and preferred side-chains
conformations.
10. Fadouloglou, V.E., Glykos*, N.M. & Kokkinidis, M. (2000), "A fast and inexpensive procedure for drying polyacrylamide gels.", Anal. Biochem., 287, 185-186.
Electronic reprint (69 KBytes) : Local copy, or directly from Anal. Biochem. 287, 185-186, Copyright © Academic Press.

 A simple procedure for drying polyacrylamide gels is described. This
only involves soaking the gel twice in low grade
ethanol : The gel is placed in a petri dish containing 5-10 gel volumes of low grade ethanol and stirred
for 10-15 minutes. After that time, the ethanol solution is refreshed and soaking continues for 5
minutes. During this second soak the gel becomes opaque, dehydrates and shrinks uniformly (without cracking)
by a factor of about 35-40%.
In the final step, the gel is removed from the ethanol solution,
placed on a hard (non-adhesive) surface, ethanol is allowed
to evaporate from its top surface, and then it is covered with a glass plate to avoid curling during
the final stages of ethanol evaporation. After few hours the gel is ready for storage.
A simple procedure for drying polyacrylamide gels is described. This
only involves soaking the gel twice in low grade
ethanol : The gel is placed in a petri dish containing 5-10 gel volumes of low grade ethanol and stirred
for 10-15 minutes. After that time, the ethanol solution is refreshed and soaking continues for 5
minutes. During this second soak the gel becomes opaque, dehydrates and shrinks uniformly (without cracking)
by a factor of about 35-40%.
In the final step, the gel is removed from the ethanol solution,
placed on a hard (non-adhesive) surface, ethanol is allowed
to evaporate from its top surface, and then it is covered with a glass plate to avoid curling during
the final stages of ethanol evaporation. After few hours the gel is ready for storage.
9. Spyridaki, A., Glykos, N.M., Kotsifaki, D., Fadouloglou, V. & Kokkinidis*, M. (2000), "Crystallization and diffraction to ultrahigh resolution (0.8A) of a designed variant of the Rop protein.", Acta Crystallogr., D56, 1015-1016.
Electronic reprint (237 KBytes) : Local copy, or directly from Acta Cryst. D56, 1015-1016, Copyright © International Union of Crystallography.

 The Rop protein is the paradigm of a highly regular 4-a-helix
bundle, and as such it has been subject to numerous structural
and mutagenesis studies. Crystals of a designed Rop variant which
establishes a continuous heptad pattern through the bend region
have been obtained by a combination of vapour diffusion and seeding
techniques. The crystals diffract to ultrahigh (0.8A)
resolution using synchrotron radiation and cryogenic conditions.
The Rop protein is the paradigm of a highly regular 4-a-helix
bundle, and as such it has been subject to numerous structural
and mutagenesis studies. Crystals of a designed Rop variant which
establishes a continuous heptad pattern through the bend region
have been obtained by a combination of vapour diffusion and seeding
techniques. The crystals diffract to ultrahigh (0.8A)
resolution using synchrotron radiation and cryogenic conditions.
8. Glykos, N.M. & Kokkinidis*, M. (2000), "On the distribution of the bulk solvent correction parameters", Acta Crystallogr., D56, 1070-1072.
Electronic reprint (384 KBytes) : Local copy, or directly from Acta Cryst. D56, 1070-1072, Copyright © International Union of Crystallography.

 The
distribution of the bulk solvent correction parameters (Bsol, ksol)
-as determined with an
exponential scaling algorithm based on Babinet's principle-
for 219 crystal
structures deposited with the Protein Data Bank is presented. The observed distribution
strongly suggests that the two parameters are not independent,
and a reasonable agreement
with the experimental data could be obtained through the application
of a simple exponential function.
Possible interpretations of this finding are discussed.
The
distribution of the bulk solvent correction parameters (Bsol, ksol)
-as determined with an
exponential scaling algorithm based on Babinet's principle-
for 219 crystal
structures deposited with the Protein Data Bank is presented. The observed distribution
strongly suggests that the two parameters are not independent,
and a reasonable agreement
with the experimental data could be obtained through the application
of a simple exponential function.
Possible interpretations of this finding are discussed.
7. Glykos*, N.M. & Kokkinidis, M. (2000), "GraphEnt : a maximum entropy program with graphics capabilities.", J. Appl. Crystallogr., 33, 982-985.
Electronic reprint (584 KBytes) : Local copy, or directly from J. Appl. Cryst. 33, 982-985, Copyright © International Union of Crystallography.

 A maximum entropy formalism aiming at the production of a
"maximally non-committal" map is a standard method in fields of
science like radioastronomy,
but a rare exception in both X-ray crystallography and electron microscopy
(or crystallography).
This is rather unfortunate, given the wealth of information that a MAXENT map can reveal,
especially when the
map itself
is the end product (for example, low resolution electron or potential density maps,
Patterson functions, deformation maps).
The program GraphEnt
attempts to automate the procedure of calculating
maximum entropy maps, with emphasis on the calculation of
difference Patterson functions
for macromolecular crystallographic problems, while
providing a useful graphical output of the current stage of the calculation.
A maximum entropy formalism aiming at the production of a
"maximally non-committal" map is a standard method in fields of
science like radioastronomy,
but a rare exception in both X-ray crystallography and electron microscopy
(or crystallography).
This is rather unfortunate, given the wealth of information that a MAXENT map can reveal,
especially when the
map itself
is the end product (for example, low resolution electron or potential density maps,
Patterson functions, deformation maps).
The program GraphEnt
attempts to automate the procedure of calculating
maximum entropy maps, with emphasis on the calculation of
difference Patterson functions
for macromolecular crystallographic problems, while
providing a useful graphical output of the current stage of the calculation.
6. Andreeva, A.E., Borissova, B.E., Mironova, R., Glykos, N.M., Kotsifaki, D., Ivanov, I., Krysteva, M. & Kokkinidis*, M. (2000), "Crystallization of type I chloramphenicol acetyltransferase : An approach based on the concept of ionic strength reducers", Acta Crystallogr., D56, 101-103.
Electronic reprint (319 KBytes) : Local copy, or directly from Acta Cryst. D56, 101-103, Copyright © International Union of Crystallography.

 Chloramphenicol acetyltransferase (CAT) is responsible for bacterial
resistance to chloramphenicol. It catalyzes inactivation of the
antibiotic by acetyl group transfer from acetyl CoA to one or
both hydroxyl groups of chloramphenicol. Type I CAT possesses
some unique properties which are not observed in other CAT variants.
Type I CAT overexpressed in E.coli was purified and crystals
with a resolution limit of 2.22 Å have been obtained
using a novel procedure which is based on the concept
of the "ionic strength reducers". The crystals have the symmetry
of spacegroup P1 and the unit cell dimensions are a=96.5, b=113.9,
c=114.2, alpha=119.9,
beta=94.1 and gamma=98.6 degrees. These
dimensions are consistent with four to six trimers per unit cell
corresponding to a solvent fraction ranging from 65 to 47%.
Chloramphenicol acetyltransferase (CAT) is responsible for bacterial
resistance to chloramphenicol. It catalyzes inactivation of the
antibiotic by acetyl group transfer from acetyl CoA to one or
both hydroxyl groups of chloramphenicol. Type I CAT possesses
some unique properties which are not observed in other CAT variants.
Type I CAT overexpressed in E.coli was purified and crystals
with a resolution limit of 2.22 Å have been obtained
using a novel procedure which is based on the concept
of the "ionic strength reducers". The crystals have the symmetry
of spacegroup P1 and the unit cell dimensions are a=96.5, b=113.9,
c=114.2, alpha=119.9,
beta=94.1 and gamma=98.6 degrees. These
dimensions are consistent with four to six trimers per unit cell
corresponding to a solvent fraction ranging from 65 to 47%.
5. Glykos*, N.M. & Kokkinidis, M. (2000), "A stochastic approach to Molecular Replacement", Acta Crystallogr., D56, 169-174.
Electronic reprint (415 KBytes) : Local copy, or directly from Acta Cryst. D56, 169-174, Copyright © International Union of Crystallography.
The CCP4 2001 Study Weekend presentation is also available for download (Powerpoint presentation, 1 MB).
The ECM 20 (Krakow, 2001) presentation is also available for download (Powerpoint presentation, 1.6 MB).
 The classical approach to the problem of placing n copies of a search
model in the asymmetric unit of a target crystal structure, is to divide
this 6n-dimensional optimisation problem into a succession of 3-dimensional
searches (rotation function followed by translation function searches for
each of the models). Here it is shown that a structure determination method
based on a reverse Monte Carlo minimisation of the conventional crystallographic
R-factor in the 6n-dimensional space defined by the rotational
and translational parameters of the n molecules, is both feasible
and practical, at least for small n. Because all parameters of all
molecules are determined simultaneously, this algorithm should improve
the signal-to-noise ratio in difficult cases involving high crystallographic/non-crystallographic
symmetry in tightly packed crystal forms. Preliminary results from the
application of this method -obtained with a space
group general computer program which has been developed for this purpose-
are presented.
The classical approach to the problem of placing n copies of a search
model in the asymmetric unit of a target crystal structure, is to divide
this 6n-dimensional optimisation problem into a succession of 3-dimensional
searches (rotation function followed by translation function searches for
each of the models). Here it is shown that a structure determination method
based on a reverse Monte Carlo minimisation of the conventional crystallographic
R-factor in the 6n-dimensional space defined by the rotational
and translational parameters of the n molecules, is both feasible
and practical, at least for small n. Because all parameters of all
molecules are determined simultaneously, this algorithm should improve
the signal-to-noise ratio in difficult cases involving high crystallographic/non-crystallographic
symmetry in tightly packed crystal forms. Preliminary results from the
application of this method -obtained with a space
group general computer program which has been developed for this purpose-
are presented.
4. Glykos, N.M. & Kokkinidis*, M. (1999), "Meaningful refinement of poly-alanine models using rigid-body simulated annealing : application to the structure determination of the A31P Rop mutant.", Acta Crystallogr., D55, 1301-1308.
Electronic reprint (1.7 MBytes) : Local copy, or directly from Acta Cryst. D55, 1301-1308, Copyright © International Union of Crystallography.
 Conventional least-squares or simulated annealing refinement of even correctly
positioned poly-alanine models of a target structure, results to a systematic
distortion of the molecular geometry and to a concomitant increase of the
mean phase difference from the correct phase set. Here it is shown that
iterative rigid-body simulated annealing refinement of poly-alanine models
employing successively fewer residues per rigid body (down to one alanine
residue per body) at a very high initial temperature (of the order of To=10000
K) and with the geometric energy terms switched on, not only preserves
the geometry of the model, but can also converge to an essentially correct
poly-alanine trace of the target structure even when the starting model
deviates systematically and significantly from the sought structure. As
an example of the application of the method we present details of the structure
determination of the alanine-31 to proline mutant of the Rop protein, where
an initial, roughly positioned poly-alanine model (giving an average phase
difference of 78.2 degrees from the final phase set) was successfully refined
against a 1.8A resolution native data set, leading to an essentially correct
model of the main chain with an average displacement of its atomic positions
from the final model of 0.275A. The phases calculated from this refined
poly-alanine model had an average difference of 43.8 degrees from the final
phase set (corresponding to a mean figure of merit of 0.72) and gave a
readily interpretable electron density map.
Conventional least-squares or simulated annealing refinement of even correctly
positioned poly-alanine models of a target structure, results to a systematic
distortion of the molecular geometry and to a concomitant increase of the
mean phase difference from the correct phase set. Here it is shown that
iterative rigid-body simulated annealing refinement of poly-alanine models
employing successively fewer residues per rigid body (down to one alanine
residue per body) at a very high initial temperature (of the order of To=10000
K) and with the geometric energy terms switched on, not only preserves
the geometry of the model, but can also converge to an essentially correct
poly-alanine trace of the target structure even when the starting model
deviates systematically and significantly from the sought structure. As
an example of the application of the method we present details of the structure
determination of the alanine-31 to proline mutant of the Rop protein, where
an initial, roughly positioned poly-alanine model (giving an average phase
difference of 78.2 degrees from the final phase set) was successfully refined
against a 1.8A resolution native data set, leading to an essentially correct
model of the main chain with an average displacement of its atomic positions
from the final model of 0.275A. The phases calculated from this refined
poly-alanine model had an average difference of 43.8 degrees from the final
phase set (corresponding to a mean figure of merit of 0.72) and gave a
readily interpretable electron density map.
3. Glykos*, N.M. (1999), "Pepinsky's Machine : an interactive, graphics-based Fourier synthesis program with applications in teaching and research.", J. Appl. Crystallogr., 32, 821-823.
Electronic reprint (660 KBytes) : Local copy, or directly from J. Appl. Cryst. 32, 821-823, FREE ITEM, Copyright © International Union of Crystallography.
 A computer program has been developed which
given a set of structure factor amplitudes for any centrosymmetric plane
group, displays the amplitude-weighted reciprocal lattice plane and allows
the user to interactively assign and modify the phases of the structure
factors, while observing the effect of these changes on the corresponding
electron density function. The program has the added feature of being able
to calculate and interactively display the electron density maps corresponding
to all phase combinations of a user-defined subset of structure factors.
The application of the program in both crystallographic teaching and research
are discussed.
A computer program has been developed which
given a set of structure factor amplitudes for any centrosymmetric plane
group, displays the amplitude-weighted reciprocal lattice plane and allows
the user to interactively assign and modify the phases of the structure
factors, while observing the effect of these changes on the corresponding
electron density function. The program has the added feature of being able
to calculate and interactively display the electron density maps corresponding
to all phase combinations of a user-defined subset of structure factors.
The application of the program in both crystallographic teaching and research
are discussed.
2. Glykos, N.M., Cesareni, G. & Kokkinidis*, M. (1999), "Protein plasticity to the extreme : Changing the topology of a 4-alpha-helical bundle with a single amino-acid substitution.", Structure 7, 597-603.
Electronic reprint (2.2 MBytes) : Local copy, or directly from ScienceDirect, Copyright © Current Biology Publications.
 The general experience from structural studies of single amino-acid substituted
mutant proteins, is that the effect of mutation is rather localised and
minor. Here we provide a counter-example to this statement by showing that
a single alanine to proline substitution in the turn region of a 4-alpha-helical
protein leads to a complete reorganisation of the whole molecule which
is converted from the canonical left-handed all-antiparallel form, to a
right-handed, mixed parallel and antiparallel bundle, which to the best
of our knowledge and belief represents a novel topological motif for this
class of proteins. Our results suggest a possible new mechanism for the
creation and evolution of protein folds, show the importance of the loop
regions in determining the allowable folding pathways and illustrate the
malleability of protein structure
(1b6q.pdb).
The general experience from structural studies of single amino-acid substituted
mutant proteins, is that the effect of mutation is rather localised and
minor. Here we provide a counter-example to this statement by showing that
a single alanine to proline substitution in the turn region of a 4-alpha-helical
protein leads to a complete reorganisation of the whole molecule which
is converted from the canonical left-handed all-antiparallel form, to a
right-handed, mixed parallel and antiparallel bundle, which to the best
of our knowledge and belief represents a novel topological motif for this
class of proteins. Our results suggest a possible new mechanism for the
creation and evolution of protein folds, show the importance of the loop
regions in determining the allowable folding pathways and illustrate the
malleability of protein structure
(1b6q.pdb).
1. Glykos, N.M., Holzenburg, A.K.H. & Phillips*, S.E.V. (1998), "Low resolution structural characterisation of the Arginine repressor/activator from Bacillus subtilis : A combined X-ray crystallographic and electron microscopical approach", Acta Crystallogr., D54, 215-225, and Acta Crystallogr., D54, 707-707.
Electronic reprint (2.6 MBytes) : Local copy, or directly from Acta Cryst. D54, 215-225, Copyright © International Union of Crystallography.
 Attempts to determine the X-ray crystal structure of the intact homohexameric arginine
repressor/activator from B. subtilis have so far been unsuccessful.
The major problem appears to be the lack of an isomorphous heavy atom derivative
with a manageable number of substitution sites. Here it is shown how electron
microscopy of thin three-dimensional crystals,
the same
as those used for the X-ray crystallographic
studies, made it possible (i) to obtain experimental support for some conclusions
drawn on the basis of X-ray data alone, (ii) to determine the low resolution
distribution of electron density in several different crystallographic
projections, and, (iii) to obtain a tentative low resolution model of the
whole hexamer.
Attempts to determine the X-ray crystal structure of the intact homohexameric arginine
repressor/activator from B. subtilis have so far been unsuccessful.
The major problem appears to be the lack of an isomorphous heavy atom derivative
with a manageable number of substitution sites. Here it is shown how electron
microscopy of thin three-dimensional crystals,
the same
as those used for the X-ray crystallographic
studies, made it possible (i) to obtain experimental support for some conclusions
drawn on the basis of X-ray data alone, (ii) to determine the low resolution
distribution of electron density in several different crystallographic
projections, and, (iii) to obtain a tentative low resolution model of the
whole hexamer.
0. Glykos, N.M. (1995), "Structural studies of the arginine repressor/activator from Bacillus subtilis", PhD thesis, Thesis advisor Prof Simon E.V. Phillips, Astbury Department of Biophysics, University of Leeds.
Electronic reprint (7.7 MBytes)
 In the presence of L-Arginine, AhrC --the Arginine-dependent
Repressor/Activator from Bacillus subtilis-- represses the
transcription of the genes encoding the anabolic and activates those
encoding the catabolic enzymes of arginine metabolism. AhrC is a homohexamer
of total molecular mass 105 kDa. It shows no homology to any of the
characterised DNA-binding motifs or DNA-binding proteins with the exception
of ArgR, the Arginine Repressor from Escherichia coli. ArgR does not
act as a transcription activator but it has been shown to be a necessary
accessory protein for the resolution --through site-specific
recombination-- of multimers of the ColE1 plasmid. Although the two
proteins share only 29% identity and are from such taxonomically distinct
prokaryotes, AhrC can complement E. coli ArgR- strains both in the
regulation of Arginine metabolism and the resolution of the ColE1 plasmid.
This thesis describes our attempts to determine the crystal structure of
AhrC. Three different crystal forms have been produced and characterised.
Useful derivatives have been prepared for two of these forms but the
determination of their heavy atom structures proved impossible. An attempt
to determine the low resolution structure of AhrC using Electron Microscopy
has been unsuccessful. Molecular Replacement using as a search model the
crystal structure of the hexameric core fragment of ArgR also failed to give
a convincing solution.
In the presence of L-Arginine, AhrC --the Arginine-dependent
Repressor/Activator from Bacillus subtilis-- represses the
transcription of the genes encoding the anabolic and activates those
encoding the catabolic enzymes of arginine metabolism. AhrC is a homohexamer
of total molecular mass 105 kDa. It shows no homology to any of the
characterised DNA-binding motifs or DNA-binding proteins with the exception
of ArgR, the Arginine Repressor from Escherichia coli. ArgR does not
act as a transcription activator but it has been shown to be a necessary
accessory protein for the resolution --through site-specific
recombination-- of multimers of the ColE1 plasmid. Although the two
proteins share only 29% identity and are from such taxonomically distinct
prokaryotes, AhrC can complement E. coli ArgR- strains both in the
regulation of Arginine metabolism and the resolution of the ColE1 plasmid.
This thesis describes our attempts to determine the crystal structure of
AhrC. Three different crystal forms have been produced and characterised.
Useful derivatives have been prepared for two of these forms but the
determination of their heavy atom structures proved impossible. An attempt
to determine the low resolution structure of AhrC using Electron Microscopy
has been unsuccessful. Molecular Replacement using as a search model the
crystal structure of the hexameric core fragment of ArgR also failed to give
a convincing solution.